Apostle technologies have been applied in many world-class R&D studies, clinical laboratory settings, and public health response and surveillance.
This page lists some of the examples in NIPT (Noninvasive Prenatal Testing). (Note: The number in front of each publication may not be sequential as it indicates the serial number in the full publication list.)
For a complete list of applications citing Apostle technologies, including publications and customer testimonials, see References.
Publications (2025)
83. Digital PCR Assay Utilizing In-Droplet Methylation-Sensitive Digestion for Estimation of Fetal cfDNA From Plasma. Richard Dannebaum, Olga Mikhaylichenko, David Siegel, Chenyu Li, Eric Hall, Severine Margeridon, Monica Herrera, Kristin Loomis, Thea Riel, Madhumita Ramesh, Maria Gencoglu, Nathan Hendel, Anthony Henriquez, Nyari Dzvova, Raymond-John Abayan, Xinhua Lin, Martin Chavez, Nazeeh Hanna. Prenatal Diagnosis, March 16, 2025 doi: https://doi.org/10.1002/pd.6774. (Download PDF)
(Note: Apostle MiniMax technology is used in this study.)
 Abstract
Abstract
Objective Recent guidelines suggest that non-invasive prenatal screening (NIPS) should be offered to all patients with singleton and twin pregnancies. Accurate determination of fetal fraction in cell-free DNA (cfDNA) is vital for reliable NIPS outcomes. We propose a methylation-based approach using droplet digital PCR (ddPCR) and methylation-sensitive restriction enzyme (MSRE) digestion for fetal fraction quantification as an affordable and fast solution.
Method Following biomarker discovery using early pregnancy placental genomic DNA (gDNA) and cfDNA from non-pregnant female individuals, we designed assays targeting MSRE-compatible regions based on contrasting methylation patterns between maternal and fetal cfDNA. We established a proof-of-concept ddPCR workflow on the Bio-Rad Droplet Digital PCR QX600 instrument.
Results Testing the fetal fraction assay multiplex on 137 prospective clinical samples demonstrated high concordance with NGS results for both female and male pregnancies as well as with chromosome Y-based calculations for samples with a male fetus. Reproducibility analysis indicated lower variability compared to previously reported NGS performance.
Conclusion This study showcases the potential of this novel, 6-color, high-multiplex methylation ddPCR panel for accurate measurement of fetal fraction in cfDNA samples. It presents opportunities to integrate such methodology as a standalone measurement to assess the quality of samples undergoing NIPS.
(Methods section)
2.2.2 | Cell‐Free DNA Sample Processing
Libraries from cfDNA were prepared using cell-free DNA extracted from 2 mL of plasma using the Apostle MiniMax High Efficiency Cell-Free DNA Isolation Kit (Apostle Inc.).
Publications (2024)
76. A novel SNP-based approach for non-invasive prenatal paternity testing using multiplex PCR targeted capture sequencing. Yiling Qu, Ranran Zhang, Li Qing, et al. Journal of Translational Genetics and Genomics. 2024;8:378-93.
(Note: Apostle MiniMax technology is used in this study.)
 Abstract
Abstract
Objective: To enhance the safety, simplicity, and efficacy of non-invasive prenatal paternity testing, we developed a method based on multiplex PCR targeted capture sequencing technology utilizing single nucleotide polymorphisms (SNPs) as genetic markers.
Method: We screened 627 SNPs from public databases and literature based on specific criteria and population genetic data from 100 unrelated individuals. A total of 15 peripheral blood samples were collected from pregnant women and the suspected father. Paternal alleles were detected and analyzed in the plasma cell-free DNA (cfDNA) of pregnant women, fetal SNP genotypes were obtained, and the combined paternity index (CPI) was calculated for paternity testing.
Results: Biological fathers were accurately determined in all cases, with CPI values ranging from 1.05 × 1014 to 2.03 × 1034, consistent with results obtained using polymerase chain reaction-capillary electrophoresis (PCR-CE) with short tandem repeats. Significant differences in CPI between unrelated males and biological fathers allowed for straightforward exclusion. Even cfDNA from maternal plasma as early as five gestational weeks enabled accurate paternity determination.
Conclusion: This novel approach demonstrates significant improvements by reducing the number of SNPs, streamlining the research procedure, and lowering costs, yielding substantial advancements in non-invasive prenatal paternity testing.
(Methods and Materials section)
Sample collection
Maternal blood samples (approximately 10 mL) were collected using MiniMax cfDNA blood collection tubes (Apostle, USA)
Publications (2023)
51. Noninvasive Prenatal Screening for Common Fetal Aneuploidies Using Single-Molecule Sequencing. Yeqing Qian, Yongfeng Liu, Kai Yan, et al. Laboratory Investigation Volume 103, Issue 4, April 2023, 100043
(Note: Apostle MiniMax technology is used in this study.)
 Amplification biases caused by next-generation sequencing (NGS) for noninvasive prenatal screening (NIPS) may be reduced using single-molecule sequencing (SMS), during which PCR is omitted. Therefore, the performance of SMS-based NIPS was evaluated. We used SMS-based NIPS to screen for common fetal aneuploidies in 477 pregnant women. The sensitivity, specificity, positive predictive value, and negative predictive value were estimated. The GC-induced bias was compared between the SMS- and NGS-based NIPS methods. Notably, a sensitivity of 100% was achieved for fetal trisomy 13 (T13), trisomy 18 (T18), and trisomy 21 (T21). The positive predictive value was 46.15% for T13, 96.77% for T18, and 99.07% for T21. The overall specificity was 100% (334/334). Compared with NGS, SMS (without PCR) had less GC bias, a better distinction between T21 or T18 and euploidies, and better diagnostic performance. Overall, our results suggest that SMS improves the performance of NIPS for common fetal aneuploidies by reducing the GC bias introduced during library preparation and sequencing.
Amplification biases caused by next-generation sequencing (NGS) for noninvasive prenatal screening (NIPS) may be reduced using single-molecule sequencing (SMS), during which PCR is omitted. Therefore, the performance of SMS-based NIPS was evaluated. We used SMS-based NIPS to screen for common fetal aneuploidies in 477 pregnant women. The sensitivity, specificity, positive predictive value, and negative predictive value were estimated. The GC-induced bias was compared between the SMS- and NGS-based NIPS methods. Notably, a sensitivity of 100% was achieved for fetal trisomy 13 (T13), trisomy 18 (T18), and trisomy 21 (T21). The positive predictive value was 46.15% for T13, 96.77% for T18, and 99.07% for T21. The overall specificity was 100% (334/334). Compared with NGS, SMS (without PCR) had less GC bias, a better distinction between T21 or T18 and euploidies, and better diagnostic performance. Overall, our results suggest that SMS improves the performance of NIPS for common fetal aneuploidies by reducing the GC bias introduced during library preparation and sequencing.
(Materials and Methods section) - cfDNA was extracted from 0.6 mL of maternal plasma using Apostle MiniMaxTM High-Efficiency cfDNA Isolation Kit (ref. number: A17622-50; Apostle) according to the manufacturer’s instructions.
Publications (2022)
40. Quantifying Fetal DNA in Maternal Blood Plasma by ddPCR Using DNA Methylation. E. Hall, T. Riel, M. Ramesh, M. Gencoglu, O. Mikhaylichenko, R. Dannebaum, S. Margeridon, M. Herrera. Bio-Rad Laboratories, Pleasanton, CA. Association for Molecular Pathology 2022 Annual Meeting Abstracts. J Mol Diagn 2022, 24:S1 Abstract G072. (Full Abstract List) (G072 Only)
(Note: Apostle MiniMax technology is used in this study.)
 Introduction: The proportion of cell-free DNA (cfDNA) circulating in maternal blood that originates from the fetus, the fetal fraction, is an important quality control metric when performing tests on fetal-derived cfDNA. Epigenetic differences produce dissimilar DNA methylation patterns, allowing for leveraging regions of high methylation contrast using methylation-sensitive restriction enzyme (MSRE) digestion to quantify fetal and maternal DNA via droplet digital PCR (ddPCR). This advancement positions ddPCR as a faster and less expensive alternative to next-generation sequencing (NGS) for fetal fraction estimation.
Introduction: The proportion of cell-free DNA (cfDNA) circulating in maternal blood that originates from the fetus, the fetal fraction, is an important quality control metric when performing tests on fetal-derived cfDNA. Epigenetic differences produce dissimilar DNA methylation patterns, allowing for leveraging regions of high methylation contrast using methylation-sensitive restriction enzyme (MSRE) digestion to quantify fetal and maternal DNA via droplet digital PCR (ddPCR). This advancement positions ddPCR as a faster and less expensive alternative to next-generation sequencing (NGS) for fetal fraction estimation.
Methods: Assays were designed to target MSRE- compatible regions with high methylation contrast between maternal and fetal cfDNA. Fetal assays targeted sites hypermethylated in fetal cfDNA and maternal assays targeted sites hypermethylated in maternal cfDNA. The assay multiplex was tested against contrived and clinical samples using an in-droplet MSRE-ddPCR workflow. The reaction mix was dropletized to create about 20,000 droplets per 24-μL reaction, thermocycled, and analyzed in a QX ONE instrument. The thermocycling profile included a 45-minute MSRE incubation step prior to PCR amplification. Contrived samples were constructed by spiking DNA-free plasma with micrococcal nuclease-digested DNA from an amniotic fluid cell line ("fetal" component) and a B-lymphocyte cell line ("maternal" component). Clinical samples were remnant diagnostic samples with existing NGS non-invasive prenatal testing results attached. DNA was extracted from all samples with the Apostle MiniMax kit on the KingFisher Flex.
Results: From an initial set of 15 assays, a final five-assay multiplex was produced following amplicon sequencing with NGS and ddPCR screening. Although amplicon sequencing did not completely predict ddPCR performance and non- specific interactions, it was valuable for guiding the final ddPCR screen. The five-assay multiplex, consisting of three fetal assays and two maternal assays, produced an excellent linear response against contrived samples from 0% to 25% fetal fraction (R2 >0.99). Similarly, a high correlation was observed between ddPCR-estimated fetal fraction and NGS fetal fraction for a set of clinical samples (n=6, plus two non- pregnant controls, R2 >0.94).
Conclusions: As an epigenetic trait, DNA methylation is a useful way to discriminate between otherwise highly similar DNA sequences in an efficient and effective manner. Leveraging DNA methylation may be done with minimal impact to the standard ddPCR workflow. The high sensitivity, speed, and direct quantification of ddPCR make it an attractive alternative to NGS for fetal fraction estimation.
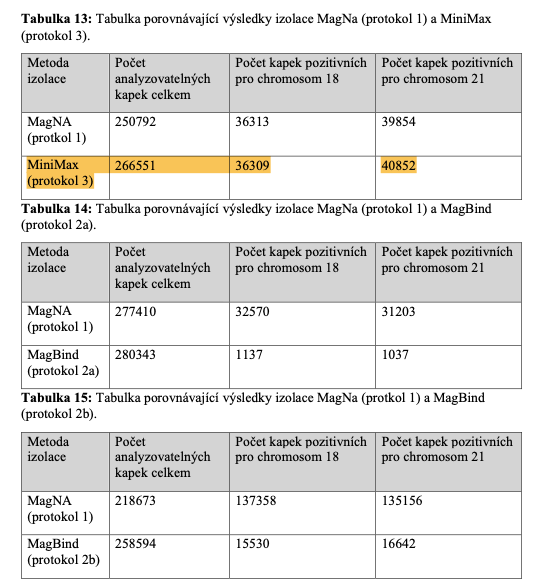
34. Optimalizace digitální polymerázové řetězové reakce pro aplikaci v neinvazivní prenatální diagnostice (Optimization of digital polymerase chain reaction for application in non-invasive prenatal diagnostics) Author: Šenkyřík, Pavel; Advisor: Korabečná, Marie; Referee: Vodička, Radek; Faculty / Institute: Faculty of Science; Discipline: Anthropology and Human Genetics; Department: Department of Anthropology and Human Genetics; Date of defense: 13. 9. 2022; Publisher: Univerzita Karlova, Přírodovědecká fakulta; Language: Czech
(Note: Apostle MiniMax technology is discussed in this diploma thesis.)
(Note: Table 13 referenced below in this diploma thesis shows that Apostle MiniMax yields the greatest number of analyzable drops in the digital PCR.)
31. A microfluidic platform for high-purity cell free DNA extraction from plasma for non-invasive prenatal testing. Lindsay Schneider, Thomas Usherwood, Anubhav Tripathi. Prenatal Diagnosis January 14, 2022; https://doi.org/10.1002/pd.6092
(Note: Apostle MiniMax technology is discussed in this study.)
 Objectives
Objectives
Increase the yield and purity of cell-free DNA (cfDNA) extracted from plasma for non-invasive prenatal testing (NIPT) as inefficiencies in this extraction and purification can dramatically affect the sensitivity and specificity of the test.
Methods
This work integrates cfDNA extraction from plasma with a microfluidic chip platform by combining magnetic bead-based extraction and electroosmotic flow on the microfluidic chip. Various wash buffers and voltage conditions were simulated using COMSOL Multiphysics Modeling and tested experimentally.
Results
When performing the first wash step of this assay on the microfluidic chip with 300 V applied across the channel there was a six-fold increase in the A260/A230 ratio showing a significant improvement (p value 0.0005) in the purity of the extracted sample all while maintaining a yield of 68.19%. These values are critical as a high yield results in more sample to analyze and an increase in A260/A230 ratio corresponds to a decrease in salt contaminants such as guanidinium thiocyanate which can interfere with downstream processes during DNA library preparation and potentially hinder the NIPT screening results.
Conclusions
This technique has the potential to improve NIPT outcomes and other clinically relevant workflows that use cfDNA as an analyte such as cancer detection.
29. Nanostructures in non-invasive prenatal genetic screening. Samira Sadeghi, Mahdi Rahaie & Bita Ostad-Hasanzadeh. Biomedical Engineering Letters volume 12, pages 3–18 (2022)
(Note: Apostle MiniEnrich technology is reviewed in this review article.)
 Abstract
Abstract
Prenatal screening is an important issue during pregnancy to ensure fetal and maternal health, as well as preventing the birth of a defective fetus and further problems such as extra costs for the family and society. The methods for the screening have progressed to non-invasive approaches over the recent years. Limitations of common standard screening tests, including invasive sampling, high risk of abortion and a big delay in result preparation have led to the introduction of new rapid and non-invasive approaches for screening. Non-invasive prenatal screening includes a wide range of procedures, including fetal cell-free DNA analysis, proteome, RNAs and other fetal biomarkers in maternal serum. These biomarkers require less invasive sampling than usual methods such as chorionic villus sampling, amniocentesis or cordocentesis. Advanced strategies including the development of nanobiosensors and the use of special nanoparticles have provided optimization and development of NIPS tests, which leads to more accurate, specific and sensitive screening tests, rapid and more reliable results and low cost, as well. This review discusses the specifications and limitations of current non-invasive prenatal screening tests and introduces a novel collection of detection methods reported studies on nanoparticles’ aided detection. It can open a new prospect for further studies and effective investigations in prenatal screening field.
(pp. 13) In another study, Zhang, et al. designed a magnetic nano-platform named MiniEnrich, used to enrich the cff-DNA fragments in maternal plasma with high resolution by developing two different magnetic nanoparticle types. It was suggested that the mentioned platform had a fair potential to help detect monogenic disorders in non-invasive prenatal screening [108].
Publications (2021)
27. Segmental duplication as potential biomarkers for non-invasive prenatal testing of aneuploidies. Xinwen Chen, Yifan Li, Qiuying Huang, Xingming Lin, Xudong Wang, Yafang Wang, Ying Liu, Qiushun He, Yinghua Liu, Ting Wang, Zhi-Liang Ji, Qingge Li. EBioMedicine August 11, 2021; https://www.thelancet.com/journals/ebiom/article/PIIS2352-3964(21)00328-5/fulltext
(Note: Apostle MiniMax technology is used in this study.)
 We developed a computational program whereby available SD regions can be processed and analyzed efficiently for their potential use as biomarkers of the aneuploidy of interest. For the five common aneuploidies, i.e., trisomy 13, 18, 21, and two sex chromosome aneuploidies, a total of 21,772 candidate SD biomarker sequences together with their corresponding primer/probe sets were generated. The primer/probe sets were tested using a real-time PCR-based multicolour melting curve analysis for simultaneous detection of the five common aneuploidies, and yielded 100% clinical sensitivity and 99.64% specificity when subjected to a clinical evaluation. Following the observations that the SD biomarkers for aneuploidy could be better detected by digital PCR with improved accuracy, we established a noninvasive prenatal testing protocol for trisomy 21 and attained 100% concordance with next generation sequencing.
We developed a computational program whereby available SD regions can be processed and analyzed efficiently for their potential use as biomarkers of the aneuploidy of interest. For the five common aneuploidies, i.e., trisomy 13, 18, 21, and two sex chromosome aneuploidies, a total of 21,772 candidate SD biomarker sequences together with their corresponding primer/probe sets were generated. The primer/probe sets were tested using a real-time PCR-based multicolour melting curve analysis for simultaneous detection of the five common aneuploidies, and yielded 100% clinical sensitivity and 99.64% specificity when subjected to a clinical evaluation. Following the observations that the SD biomarkers for aneuploidy could be better detected by digital PCR with improved accuracy, we established a noninvasive prenatal testing protocol for trisomy 21 and attained 100% concordance with next generation sequencing.
Our study confirmed that SD regions are preferred biomarkers for aneuploidy detection and in particular SD-based digital PCR could find potential use for NIPT of trisomy. A similar strategy can be applied to other chromosomal abnormality and genetic disorders.
Publications (2020)
18. High-resolution DNA size enrichment using a magnetic nano-platform and application in non-invasive prenatal testing. Bo Zhang, Shuting Zhao, Hao Wan, Ying Liu, Fei Zhang, Xin Guo, Wenqi Zeng, Haiyan Zhang, Linghua Zeng, Jiale Qu, Ben-Qing Wu, Xinhong Wan, Charles R. Cantor and Dongliang Ge. Analyst. July 2020, 145, 5733-5739
 Precise DNA sizing can boost sequencing efficiency, reduce cost, improve data quality, and even allow sequencing of low-input samples, while current pervasive DNA sizing approaches are incapable of differentiating DNA fragments under 200 bp with high resolution (<20 bp). In non-invasive prenatal testing (NIPT), the size distribution of cell-free fetal DNA in maternal plasma (main peak at 143 bp) is significantly different from that of maternal cell-free DNA (main peak at 166 bp). The current pervasive workflow of NIPT and DNA sizing is unable to take advantage of this 20 bp difference, resulting in sample rejection, test inaccuracy, and restricted clinical utility. Here we report a simple, automatable, high-resolution DNA size enrichment workflow, named MiniEnrich, on a magnetic nano-platform to exploit this 20 bp size difference and to enrich fetal DNA fragments from maternal blood. Two types of magnetic nanoparticles were developed, with one able to filter high-molecular-weight DNA with high resolution and the other able to recover the remaining DNA fragments under the size threshold of interest with >95% yield. Using this method, the average fetal fraction was increased from 13% to 20% after the enrichment, as measured by plasma DNA sequencing. This approach provides a new tool for high-resolution DNA size enrichment under 200 bp, which may improve NIPT accuracy by rescuing rejected non-reportable clinical samples, and enable NIPT earlier in pregnancy. It also has the potential to improve non-invasive screening for fetal monogenic disorders, differentiate tumor-related DNA in liquid biopsy and find more applications in autoimmune disease diagnosis.Two types of magnetic nanoparticles were developed, with one able to filter high-molecular-weight DNA with high resolution and the other able to recover the remaining DNA fragments under the size threshold of interest with >95% yield. Using this method, the average fetal fraction was increased from 13% to 20% after the enrichment, as measured by plasma DNA sequencing. This approach provides a new tool for high-resolution DNA size enrichment under 200 bp, which may improve NIPT accuracy by rescuing rejected non-reportable clinical samples, and enable NIPT earlier in pregnancy. It also has the potential to improve non-invasive screening for fetal monogenic disorders, differentiate tumor-related DNA in liquid biopsy and find more applications in autoimmune disease diagnosis.
Precise DNA sizing can boost sequencing efficiency, reduce cost, improve data quality, and even allow sequencing of low-input samples, while current pervasive DNA sizing approaches are incapable of differentiating DNA fragments under 200 bp with high resolution (<20 bp). In non-invasive prenatal testing (NIPT), the size distribution of cell-free fetal DNA in maternal plasma (main peak at 143 bp) is significantly different from that of maternal cell-free DNA (main peak at 166 bp). The current pervasive workflow of NIPT and DNA sizing is unable to take advantage of this 20 bp difference, resulting in sample rejection, test inaccuracy, and restricted clinical utility. Here we report a simple, automatable, high-resolution DNA size enrichment workflow, named MiniEnrich, on a magnetic nano-platform to exploit this 20 bp size difference and to enrich fetal DNA fragments from maternal blood. Two types of magnetic nanoparticles were developed, with one able to filter high-molecular-weight DNA with high resolution and the other able to recover the remaining DNA fragments under the size threshold of interest with >95% yield. Using this method, the average fetal fraction was increased from 13% to 20% after the enrichment, as measured by plasma DNA sequencing. This approach provides a new tool for high-resolution DNA size enrichment under 200 bp, which may improve NIPT accuracy by rescuing rejected non-reportable clinical samples, and enable NIPT earlier in pregnancy. It also has the potential to improve non-invasive screening for fetal monogenic disorders, differentiate tumor-related DNA in liquid biopsy and find more applications in autoimmune disease diagnosis.Two types of magnetic nanoparticles were developed, with one able to filter high-molecular-weight DNA with high resolution and the other able to recover the remaining DNA fragments under the size threshold of interest with >95% yield. Using this method, the average fetal fraction was increased from 13% to 20% after the enrichment, as measured by plasma DNA sequencing. This approach provides a new tool for high-resolution DNA size enrichment under 200 bp, which may improve NIPT accuracy by rescuing rejected non-reportable clinical samples, and enable NIPT earlier in pregnancy. It also has the potential to improve non-invasive screening for fetal monogenic disorders, differentiate tumor-related DNA in liquid biopsy and find more applications in autoimmune disease diagnosis.
For a complete list of publications citing Apostle technologies, see Publications.