Background
DNA methylation, an epigenetic modification, is closely related to the gene expression and chromatin remodeling. Abnormal methylation status is closely related to the occurrence and development of various diseases such as cancer. The precise identification of cytosine methylation status in genome plays a critical role in research and clinical areas, especially in precise oncology. In terms of analyzing methylation status, with the treatment of bisulfite or enzymes, the unmethylated cytosine can be converted to uracil, whereas the methylated cytosine remains unchanged. Afterwards, combined with high-throughput sequencing technology, the genomic methylation status can be analyzed massively with single nucleotide resolution. However, there are still cost and efficiency limitations for the analysis of whole genomes, the introduction of targeted enrichment technology can reduce the sequencing cost and improve the targeted analysis efficiency by improving the sequencing depth of interested regions. Apostle offers an NGS total solution for capture-based methylation sequencing (Methyl-Seq) on Illumina sequencing platform, including Methyl-Seq library preparation kit based on bisulfite or enzymatic conversion, methylation bisulfite conversion module, design & synthesis service of capture probes, hybridization reagents related and integrated analysis tools. Designed to help researchers quickly and accurately analyze the methylation status of the genome and provide support for research and clinical diagnosis of related diseases.
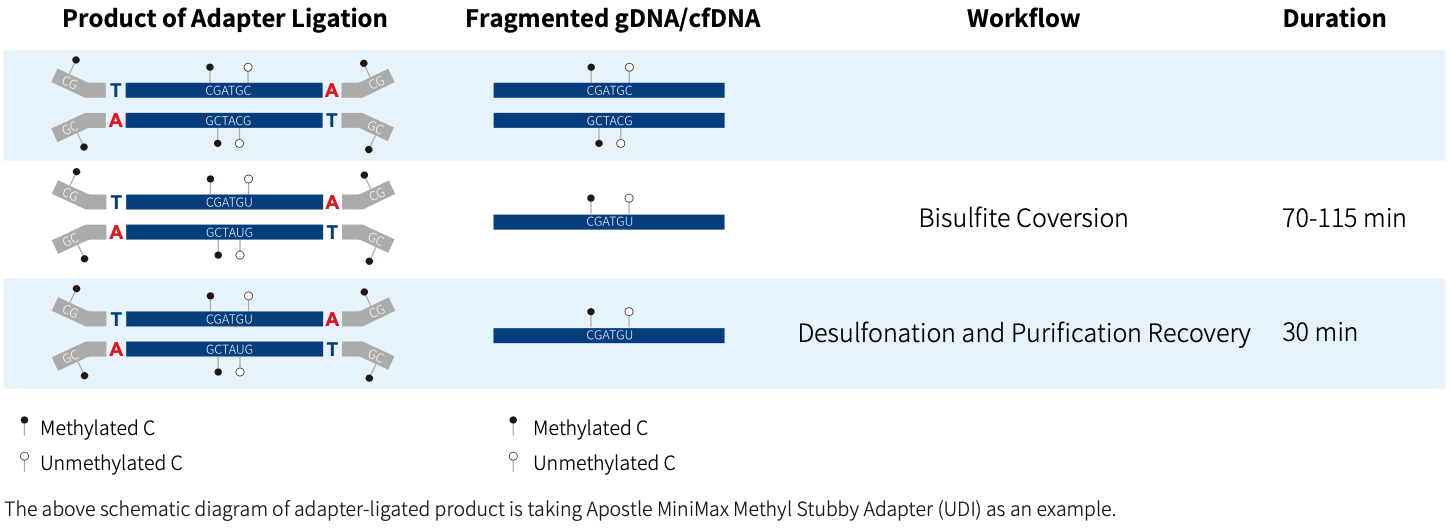
Workflow
|
|
Introduction
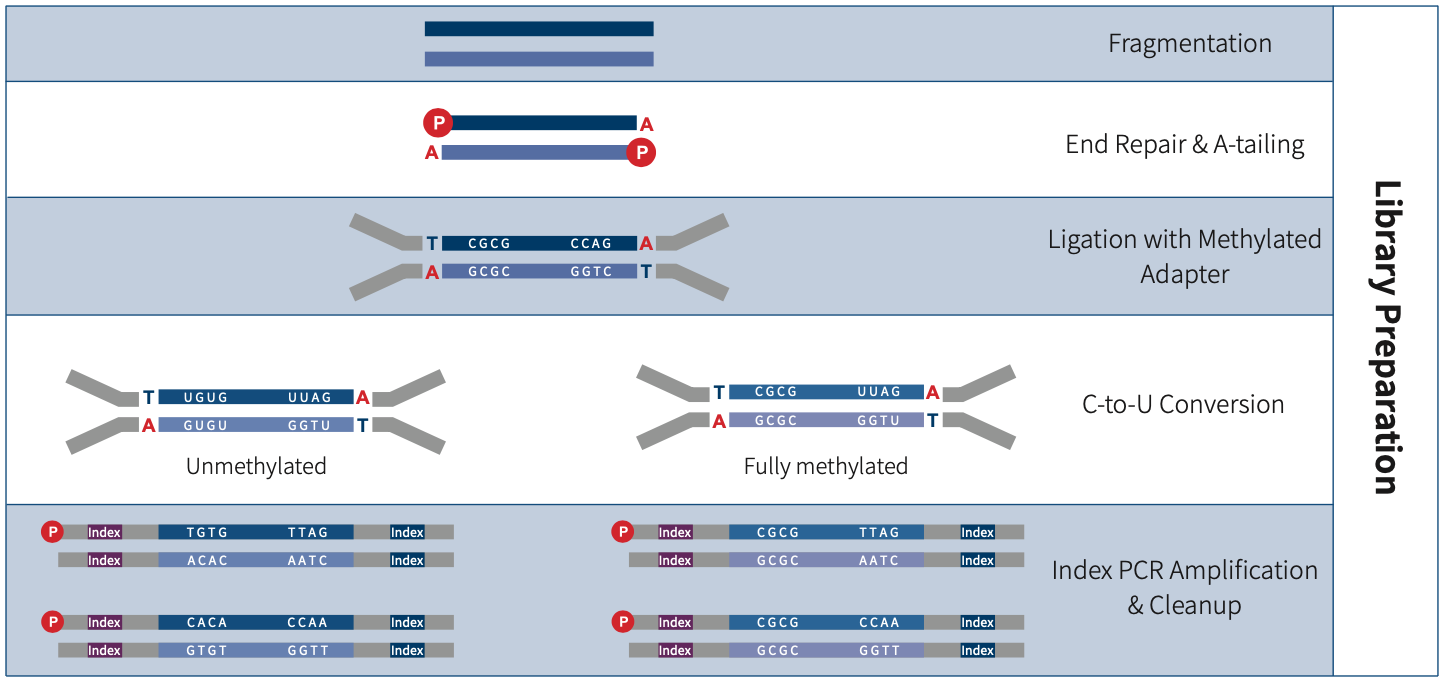
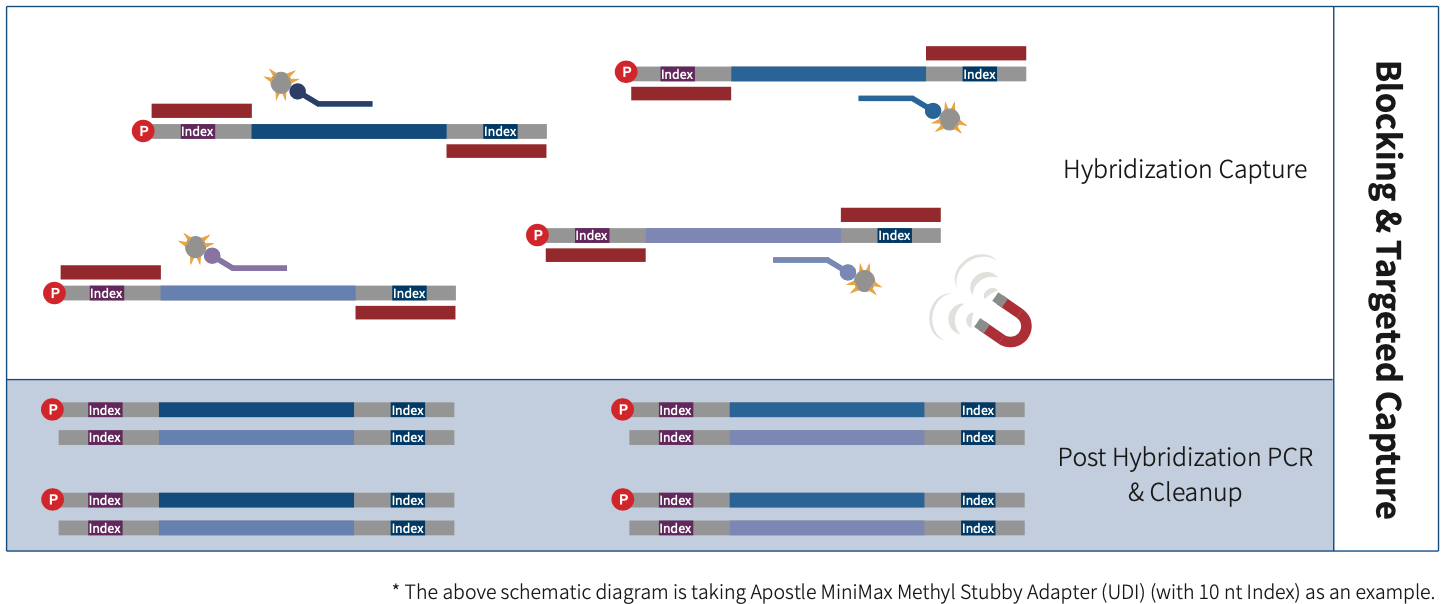
Apostle MiniMax Methyl Library Preparation Module is a double-strand DNA methylation sequencing library preparation kit developed for MGI & Illumina sequencing platforms. It supports multiple options for adapters selection, including Vendor N Methyl Adapter (SI) Module (for MGI), Vendor N Methyl Adapter (MDI) Module (for MGI) and Apostle MiniMax Methyl Stubby Adapter (UDI) Module (with 10 nt Index), etc. This kit can be used to prepare whole-genome methylation sequencing library. When combined with Apostle MiniMax's liquid-phase hybridization capture series products, it enables targeted enrichment of methylation sequencing library.
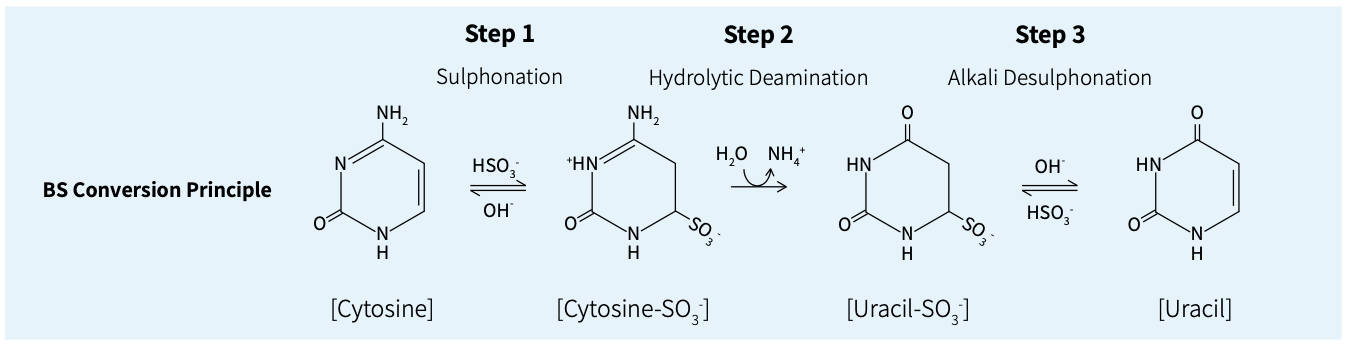
Apostle MiniMax DNA Methyl Bisulfite Conversion Module is a methylation conversion reagent developed for DNA samples based on the principle of bisulfite conversion. The conversion module, based on a magnetic bead purification protocol, efficiently converts unmethylated cytosine to uracil within 2 hr, leaving methylated cytosine unchanged, and it can be compatible with automated workstations. It can be seamlessly integrated with Apostle's double-strand methylation library preparation scheme and single-strand methylation library preparation scheme, allowing flexible selection among various types of starting samples.
Bisulfite conversion workflow
|
|
Feature
o Compatible with multiple types of samples and supports a wide range of input amounts
o Different types of adapter modules can be selected for matching according to the sequencing platform
o Efficient and stable cytosine conversion rate
o Excellent detection sensitivity of 5mC and 5hmC
o Convenient operation is conducive to automation implementation
Performance
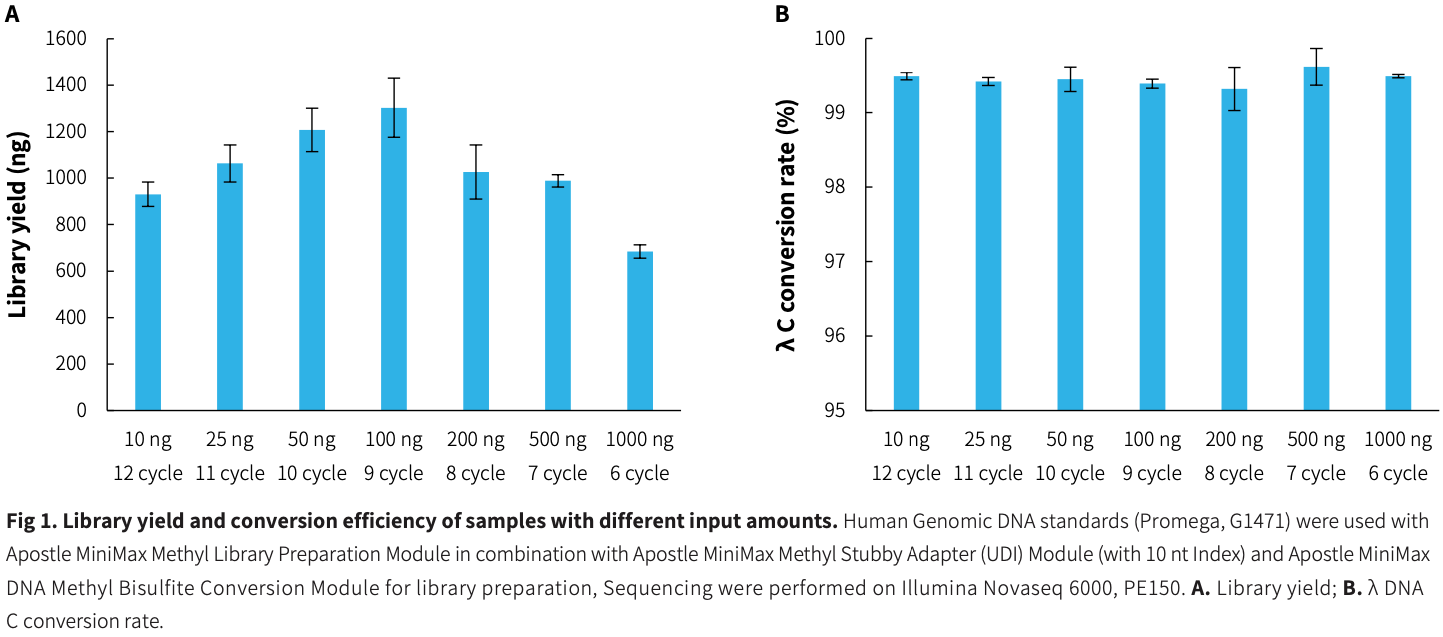
Library yield and conversion efficiency
|
|
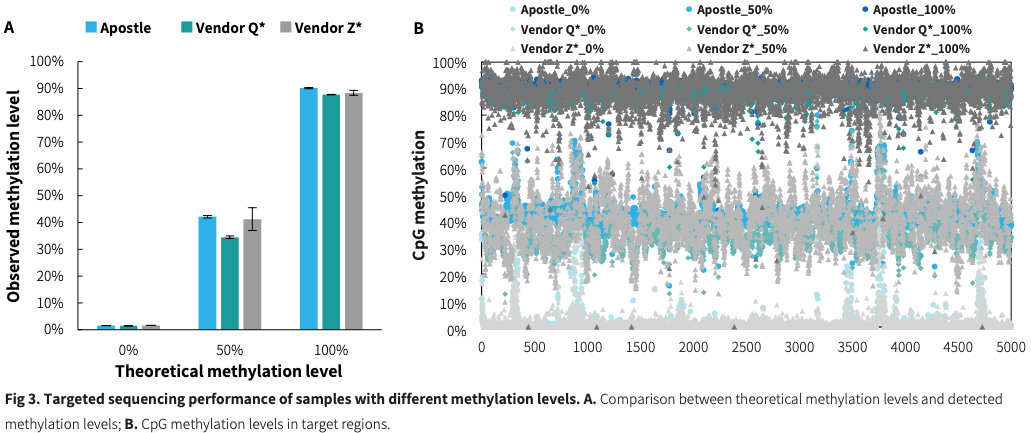
Methylation detection
|
|
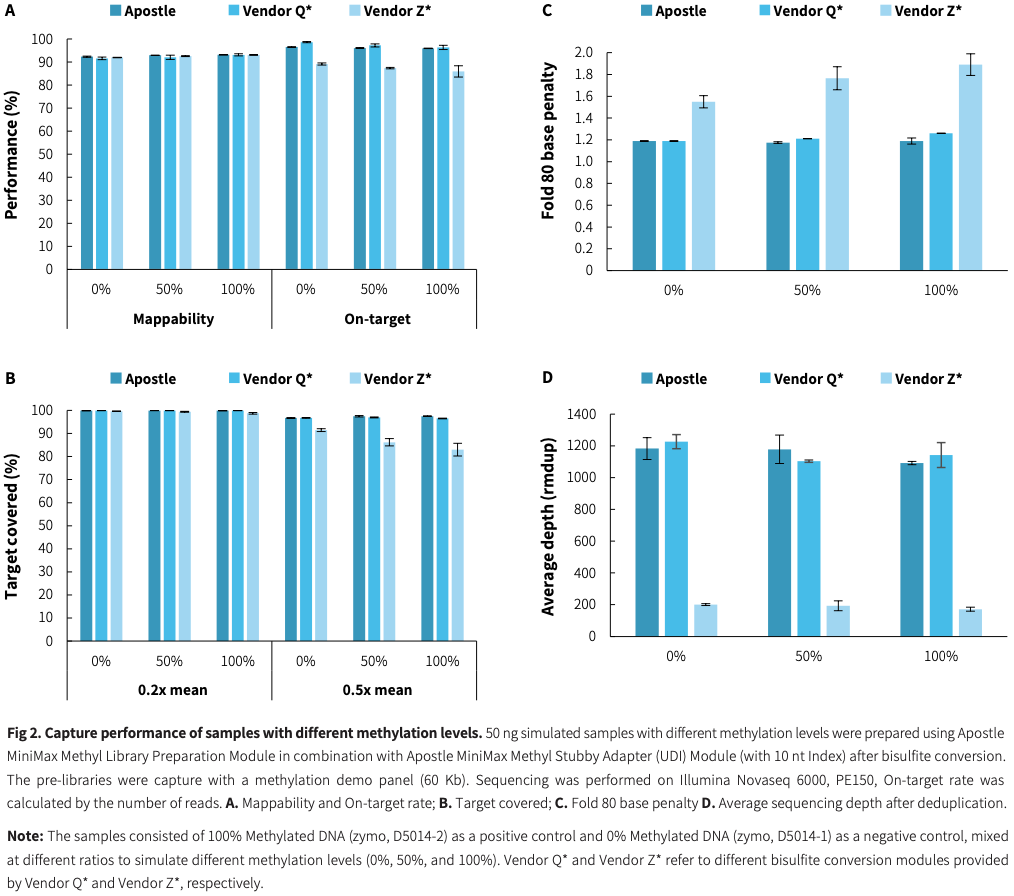
Capture performance
|
|