Background
With continuous innovation of the gene sequencing-related technologies and expanding of its application fields, the industry competitive landscape is evolving. The MGI sequencing platform consistently generates high-quality sequencing data with lower duplication rate compared to the Illumina platform, and the index hopping issue is greatly alleviated. Currently, increasing numbers of users have both Illumina and MGI sequencers at the same time. Can the same set of libraries be used for sequencing on both Illumina and MGI platforms in order to maximize the productivity? To solve this problem, MGI has launched MGIEasy Universal Library Conversion Kit (App-A) to convert Illumina platform libraries into single-stranded circular libraries, which allows for sequencing on MGI platform. However, this approach requires an additional 5-10 cycles of PCR amplification, which may introduce amplification errors and eliminates the low duplication advantage of the MGI platform. In addition, Illumina platform libraries include mostly pair-end 8 nt Index, and there are some incompatibilities in the reading of index after transformation.
Therefore, Apostle has launched an Apostle MiniMax Universal Stubby Adapter (UDI) Module. With its newly designed 10 nt Unique Dual Index, the module supports 8 nt or 10 nt Index reading mode on the Illumina platform and is also compatible with MGI sequencing platform. The libraries can be sequenced directly on MGI platform after circularization without conversion.
Solution
|
|
Introduction
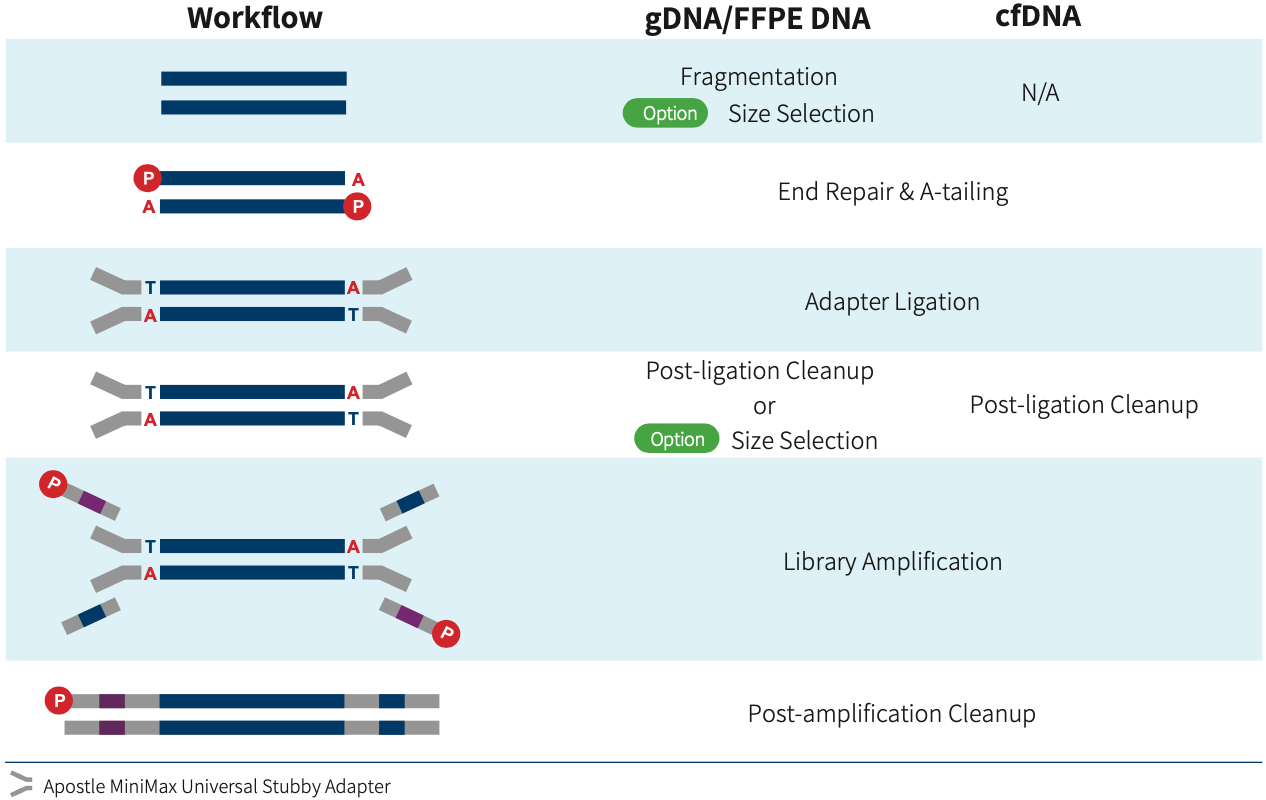
The Apostle MiniMax Universal Stubby Adapter (UDI) Module contains universal stubby adapters and 10 nt Unique Dual Index amplification primers. It can be used for preparing libraries with Illumina structures and 5' phosphorylation modification; the libraries can be sequenced on Illumina platform directly and on MGI platform after circularization without App-A conversion. The Apostle MiniMax Universal Stubby Adapter (UDI) Module is designed to be 4-Index balanced. It means the minimum balanced group only requires 4 libraries, either on the Illumina platform or MGI platform.
Performance
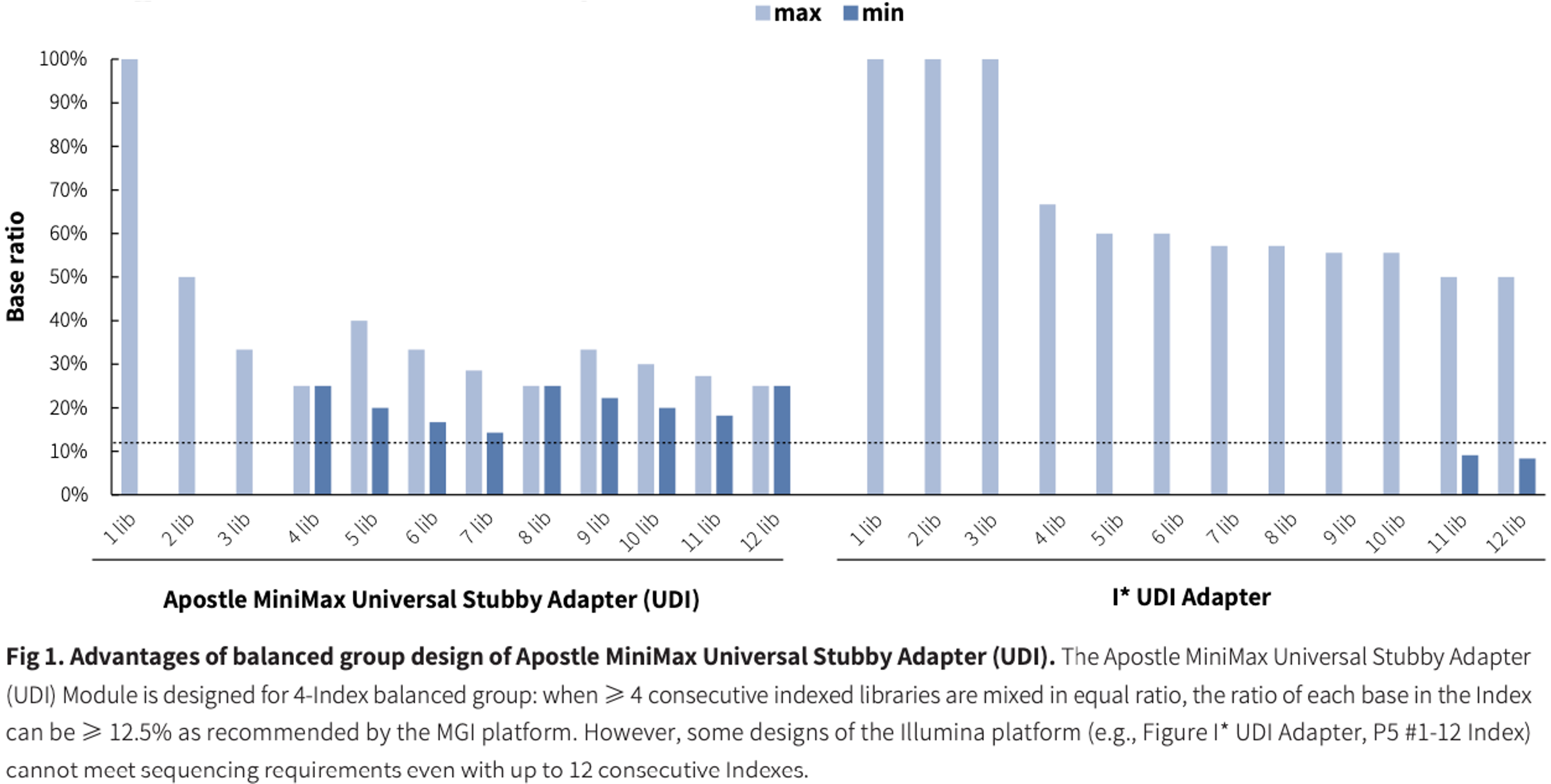
Advantage of 4-Index Balanced Group
|
|
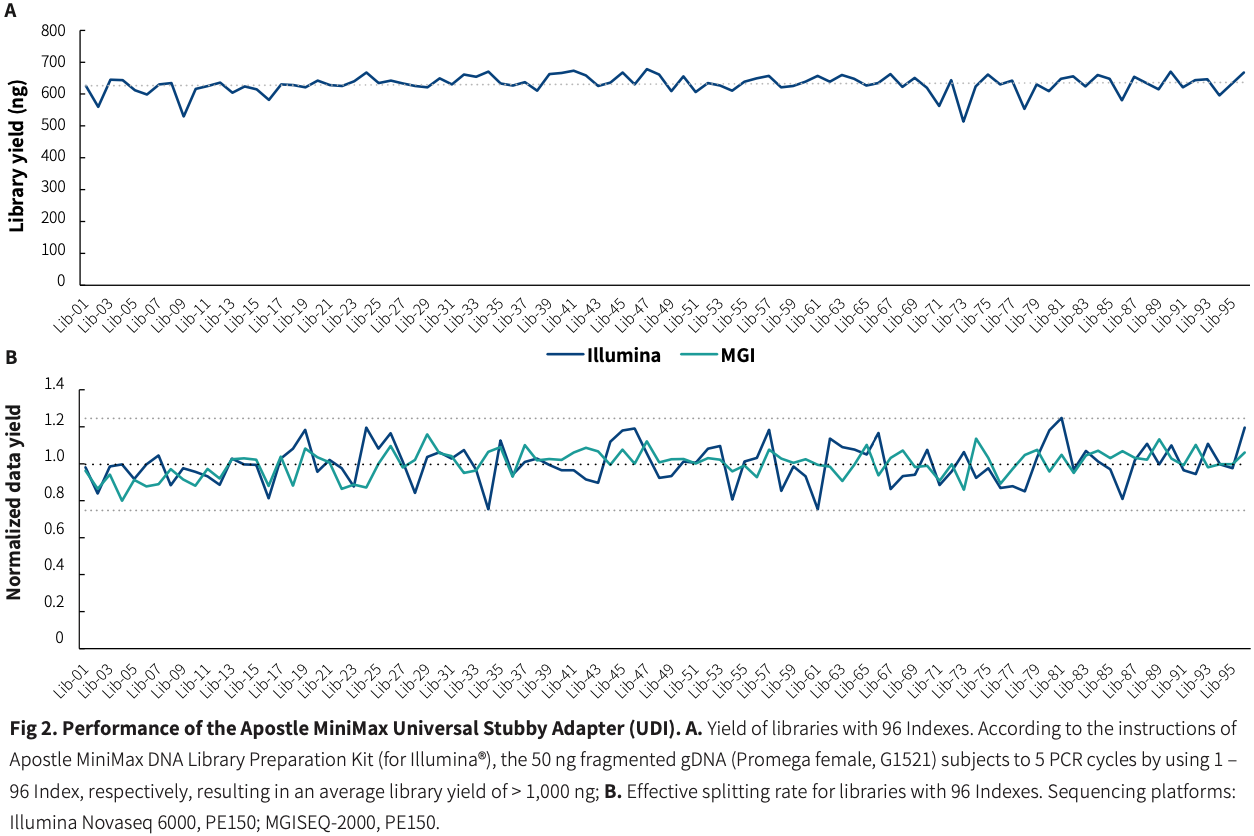
Uniform Data Output
|
|
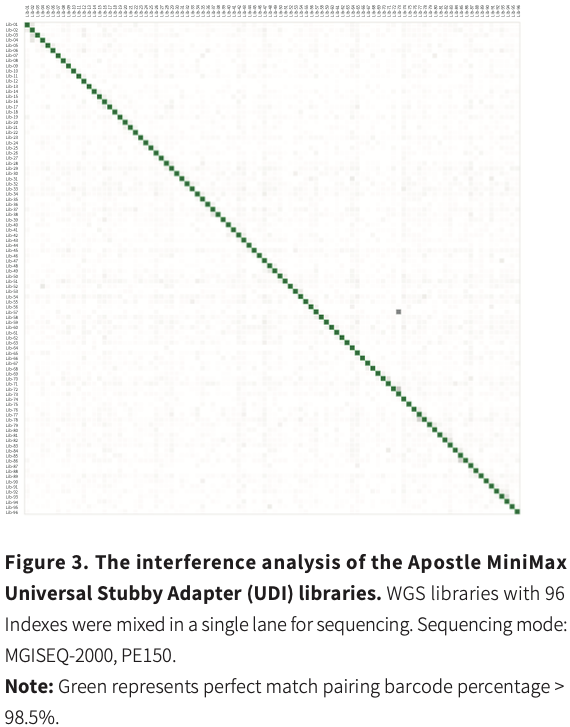
Effectively Improve Data Accuracy
|
|
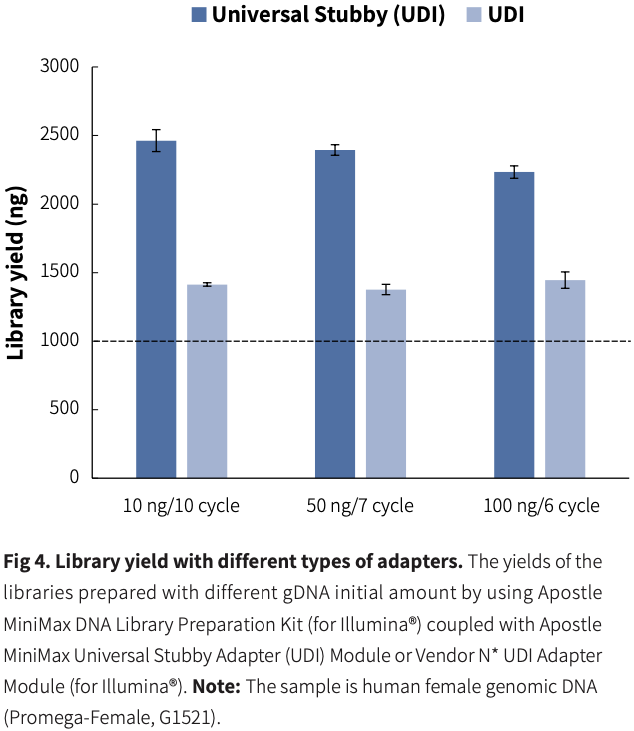
High Efficiency and Stability
|
|