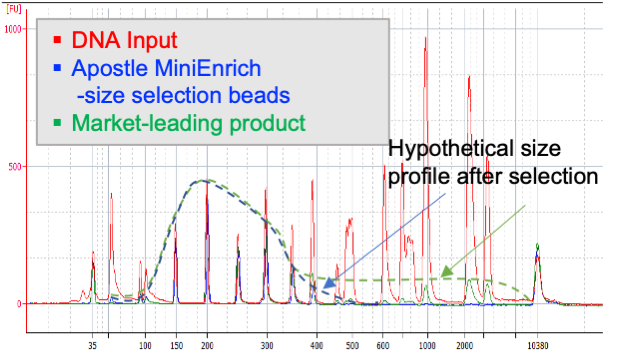
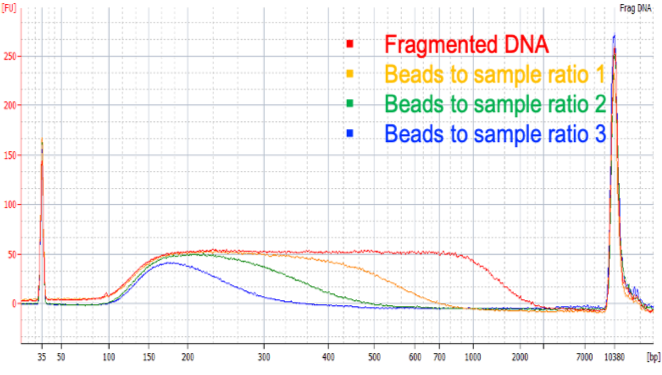
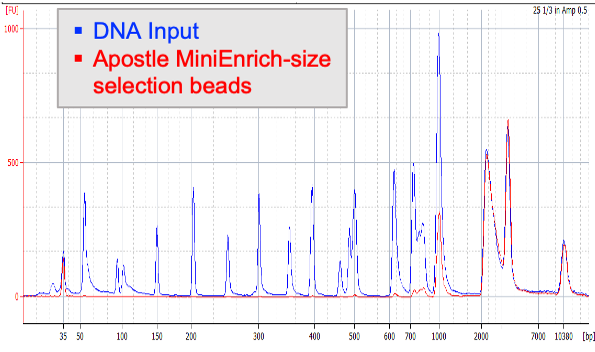
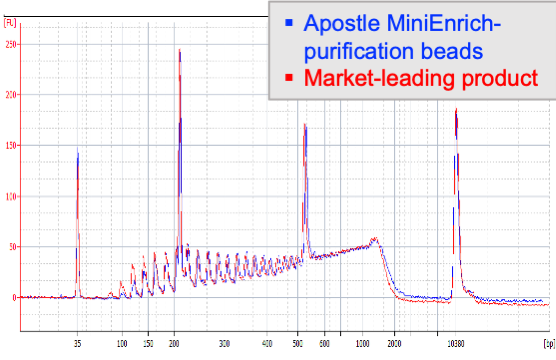
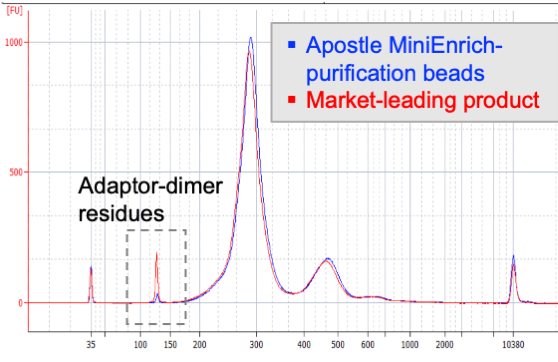
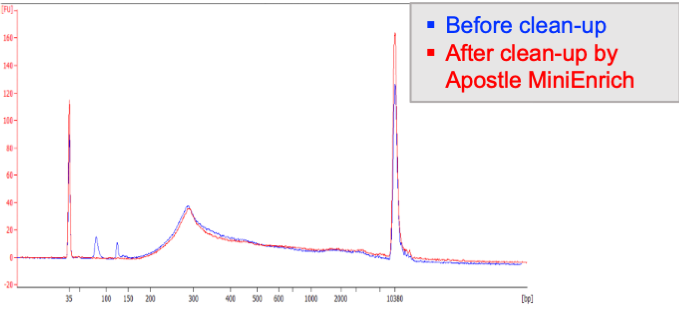
Next-generation purification, size selection, and long-read size selection
-
Promotion
Special Promotion
-
Products
Sectors
Categories
Product Information
- How to order
- [Catalog] 2024 May (PDF)
- [Flyer] MiniMax: Best-in-class gDNA/cfDNA Isolation Technology (PDF)
- [Flyer] MagTouch 3000: World 1st Large-Volume Fully-Automated cfDNA Extractor (PDF)
- [Flyer] Best-in-Class total-RNA/cfRNA Isolation Technology (PDF)
- [Flyer] Cost-efficient & High-quality Laboratory Consumables (PDF)
- [Flyer] RNA Isolation Solution (PDF)
-
Technology
- Apostle MiniMax Technology
- Apostle MiniMax (cfRNA)
- Apostle MagTouch Technology
- Apostle BCT Technology
- Apostle MiniEnrich Technology
- Apostle MiniGenomics Technology
- Apostle Triton Technology
- Apostle MiniMax (Type S)
- Apostle Viral RNA/DNA Isolation Automation System
- Apostle Well Plates and Accessories for Nucleic Acids Isolation
- Applications
- Sign In