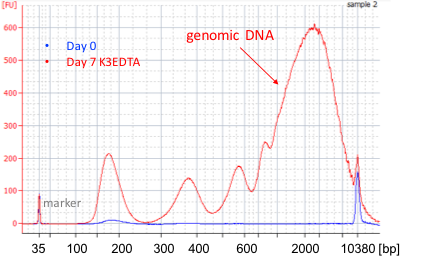
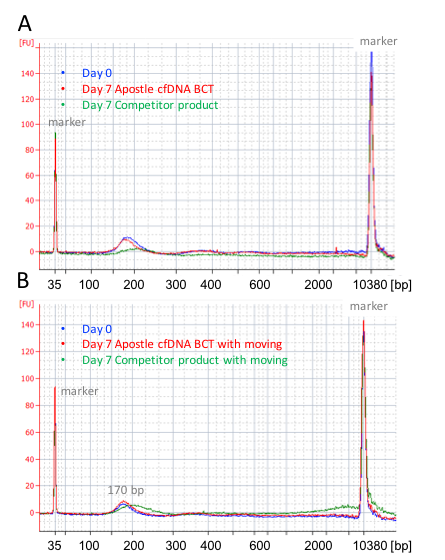
Apostle cfDNA BCT minimizes genomic DNA release during blood storage (Exhibit 2A) and transportation (Exhibit 2B) for at least 7 days. Minimum genomic DNA contamination leads to accurate and sensitive cfDNA analysis. Though competitor product also has the ability to prevent genomic DNA release, its chemistry causes crosslinking of DNA to other biomolecules, leading to size increase and a peak shift from 170bp to 200bp (Exhibit 2). This crosslinking caused by competitor’s product will affect subsequent application of isolated cfDNA. On the other hand, cfDNA preserved with Apostle cfDNA BCT for 7 days shows no peak shift compared with cfDNA isolated immediately after blood collection (Exhibit 2), demonstrating high cfDNA quality preserved by Apostle cfDNA BCT.
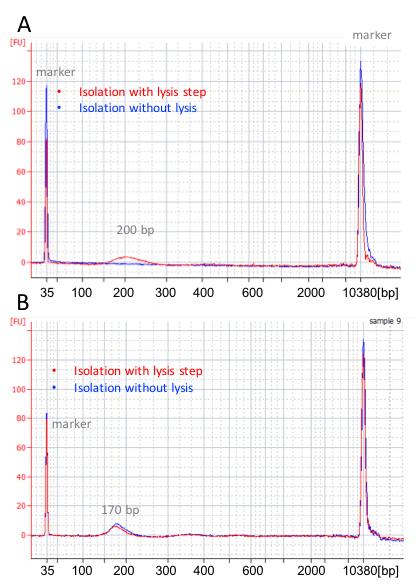
During cfDNA isolation process, according to various cfDNA isolation protocols, the proteinase K treatment of plasma can always be skipped for blood collected in EDTA tubes. However, proteinase K treatment is required for blood collected in competitor’s cfDNA blood collection tube, or no cfDNA can be isolated (Exhibit 3A). This is another evidence showing the crosslinking of cfDNA with other biomolecules during blood storage in competitor’s blood collection tube. On the other hands, cfDNA isolated from blood collected in Apostle cfDNA BCT does not require proteinase K treatment, and has the right peak at ~170 bp (Exhibit 3B), demonstrating minimal crosslinking for cfDNA preserved with Apostle cfDNA BCT.
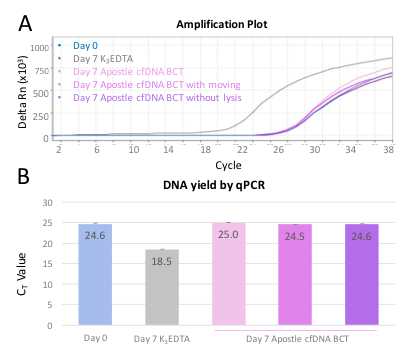
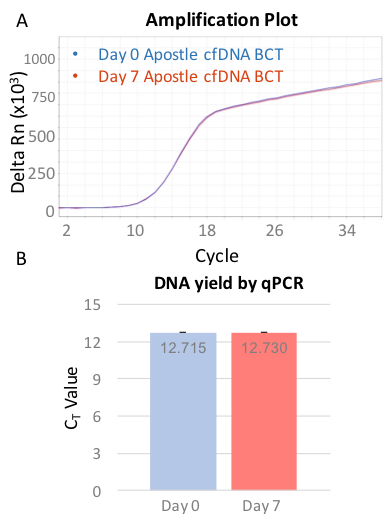
qPCR measurement for beta-globin expression of isolated cfDNA were also performed as a mean to reflect cfDNA preservation in Apostle cfDNA BCT. After 7 days storage or transportation, beta-globin expression of isolated cfDNA share the same CT value with cfDNA isolated immediately after blood drawn in EDTA tubes (Exhibit 4), demonstrating high endogenous cfDNA recovery and quality of blood collected in Apostle cfDNA BCT.
Spike in and recover experiment was performed to further validate cfDNA recovery rate of blood stored in Apostle cfDNA BCT. DNA fragment containing the EGFR c.2573T>G L858R mutation (synthetic, ~170 bp) was spiked into blood in Apostle cfDNA BCTs, and recovered after 7 days storage. qPCR measurement for EGFR c.2573T>G L858R mutation expression showed no obvious difference after 7 days storage. According to CT value, 99% of spiked in DNA was recovered, demonstrating high stability of DNA preserved with Apostle cfDNA BCT (Exhibit 5).
In summary, high cfDNA quality can be preserved with Apostle cfDNA Blood Collection Tube for downstream applications, through the prevention of cfDNA degradation, crosslingking and contamination.