What our community say
"These data establish that tumor and cfDNA methylation can be used to identify SCLC subtypes and might guide precision SCLC therapy. "..."(Methods section) cfDNA was extracted using the Apostle MiniMax High Efficiency Cell-Free DNA Isolation Kit (Apostle Inc). " ..."(Key Resources Table - Critical commercial assays) Apostle MiniMax High Efficiency Cell-Free DNA Isolation Kit. Apostle Bio A17622-250 "
"We perform transcriptome-wide characterizations in 165 lung cancer, 30 breast cancer, 37 colorectal cancer, 55 gastric cancer, 15 liver cancer, and 133 cancer-free participants and demonstrate its ability to identify transcriptomic changes occurring in early-stage tumors. "..."(Methods section) 200 μl of plasma samples were subjected to cfRNA extraction using the Apostle MiniMax High-Efficiency cfRNA Isolation Kit (Apostle), following the manufacturer’s protocol with minor modifications. "
"Our integrative model detects early-stage cancers, including those of pancreatic origin, with high sensitivity that is comparable to that of late-stage detection. "..."(Methods section) cfDNA was extracted from 0.4 mL plasma ... and eluted in a final volume of 22 μL, using an Apostle MiniMax High Efficiency cfDNA Isolation Kit (Apostle, US) according to the manufacturer’s instructions. "
"Exploratory biomarker analyses showed decreased circulating tumor DNA (ctDNA) in patients with prolonged OS... cfDNA was extracted from the entire plasma volume of a single draw using the Apostle MiniMax cfDNA Isolation kit (ApostleBio) "
"The model detected four of the five HCC cases in the cohort, showing 80% sensitivity and 94% specificity. These findings demonstrate that the MCP technology has potential for the discovery and validation of multiomics biomarkers for the noninvasive detection of cancer. This study also provides a comprehensive database of genetic and epigenetic alterations in the cfDNA of a large cohort of HCC cases and high-risk non-HCC individuals." "cfDNA was extracted from the plasma samples using the Apostle MiniMax cfDNA isolation kit (C43468, Apostle). "
Featured Publications
Cancer Cell (IF=50.3) 2024
- Tumor- and circulating-free DNA methylation identifies clinically relevant small cell lung cancer subtypes. Simon Heeke, Carl M. Gay, Marcos R. Estecio, et al. Cancer Cell January 25, 2024
(Note: Apostle MiniMax technology is used in this study.)
Abstract Small cell lung cancer (SCLC) is an aggressive malignancy composed of distinct transcriptional subtypes, but implementing subtyping in the clinic has remained challenging, particularly due to limited tissue availability. Given the known epigenetic regulation of critical SCLC transcriptional programs, we hypothesized that subtype-specific patterns of DNA methylation could be detected in tumor or blood from SCLC patients. Using genomic-wide reduced-representation bisulfite sequencing (RRBS) in two cohorts totaling 179 SCLC patients and using machine learning approaches, we report a highly accurate DNA methylation-based classifier (SCLC-DMC) that can distinguish SCLC subtypes. We further adjust the classifier for circulating-free DNA (cfDNA) to subtype SCLC from plasma. Using the cfDNA classifier (cfDMC), we demonstrate that SCLC phenotypes can evolve during disease progression, highlighting the need for longitudinal tracking of SCLC during clinical treatment. These data establish that tumor and cfDNA methylation can be used to identify SCLC subtypes and might guide precision SCLC therapy.
(Methods section)
Critical commercial assays
Apostle MiniMax High Efficiency Cell-Free DNA Isolation Kit Apostle Bio A17622-250
Nucleic acid extraction
cfDNA was extracted using the Apostle MiniMax High Efficiency Cell-Free DNA Isolation Kit (Apostle Inc).
Nature Communications 2024
- Terminal modifications independent cell-free RNA sequencing enables sensitive early cancer detection and classification. Jun Wang, Jinyong Huang, Yunlong Hu, et al. Nature Communications 15, Article number: 156 (2024)
(Note: Apostle MiniMax technology is used in this study.)
Abstract Cell-free RNAs (cfRNAs) offer an opportunity to detect diseases from a transcriptomic perspective, however, existing techniques have fallen short in generating a comprehensive cell-free transcriptome profile. We develop a sensitive library preparation method that is robust down to 100 µl input plasma to analyze cfRNAs independent of their 5’-end modifications. We show that it outperforms adapter ligation-based method in detecting a greater number of cfRNA species. We perform transcriptome-wide characterizations in 165 lung cancer, 30 breast cancer, 37 colorectal cancer, 55 gastric cancer, 15 liver cancer, and 133 cancer-free participants and demonstrate its ability to identify transcriptomic changes occurring in early-stage tumors. We also leverage machine learning analyses on the differentially expressed cfRNA signatures and reveal their robust performance in cancer detection and classification. Our work sets the stage for in-depth study of the cfRNA repertoire and highlights the value of cfRNAs as cancer biomarkers in clinical applications.
(Methods section) cfRNA extraction
Frozen plasma samples were thawed on ice prior to cfRNA extraction. 200 μl of plasma samples were subjected to cfRNA extraction using the Apostle MiniMax High-Efficiency cfRNA Isolation Kit (Apostle), following the manufacturer’s protocol with minor modifications.
Nature Communications 2023
- Integrative modeling of tumor genomes and epigenomes for enhanced cancer diagnosis by cell-free DNA. Mingyun Bae, Gyuhee Kim, Tae-Rim Lee, et al. Nature Communications 14, Article number: 2017 (2023)
(Note: Apostle MiniMax technology is used in this study.)
Abstract Multi-cancer early detection remains a key challenge in cell-free DNA (cfDNA)-based liquid biopsy. Here, we perform cfDNA whole-genome sequencing to generate two test datasets covering 2125 patient samples of 9 cancer types and 1241 normal control samples, and also a reference dataset for background variant filtering based on 20,529 low-depth healthy samples. An external cfDNA dataset consisting of 208 cancer and 214 normal control samples is used for additional evaluation. Accuracy for cancer detection and tissue-of-origin localization is achieved using our algorithm, which incorporates cancer type-specific profiles of mutation distribution and chromatin organization in tumor tissues as model references. Our integrative model detects early-stage cancers, including those of pancreatic origin, with high sensitivity that is comparable to that of late-stage detection. Model interpretation reveals the contribution of cancer type-specific genomic and epigenomic features. Our methodologies may lay the groundwork for accurate cfDNA-based cancer diagnosis, especially at early stages..
(Methods section) cfDNA was extracted from 0.4 mL plasma ... and eluted in a final volume of 22 μL, using an Apostle MiniMax High Efficiency cfDNA Isolation Kit (Apostle, US) according to the manufacturer’s instructions.
Nature Medicine 2022
- Individualized, heterologous chimpanzee adenovirus and self-amplifying mRNA neoantigen vaccine for advanced metastatic solid tumors: phase 1 trial interim results. Christine D. Palmer, Amy R. Rappaport, Matthew J. Davis, et al. Nature Medicine volume 28, pages 1619–1629 (2022)
(Note: Apostle MiniMax technology is used in this study.)
Abstract Checkpoint inhibitor (CPI) therapies provide limited benefit to patients with tumors of low immune reactivity. T cell-inducing vaccines hold promise to exert long-lasting disease control in combination with CPI therapy. Safety, tolerability and recommended phase 2 dose (RP2D) of an individualized, heterologous chimpanzee adenovirus (ChAd68) and self-amplifying mRNA (samRNA)-based neoantigen vaccine in combination with nivolumab and ipilimumab were assessed as primary endpoints in an ongoing phase 1/2 study in patients with advanced metastatic solid tumors (NCT03639714). The individualized vaccine regimen was safe and well tolerated, with no dose-limiting toxicities. Treatment-related adverse events (TRAEs) >10% included pyrexia, fatigue, musculoskeletal and injection site pain and diarrhea. Serious TRAEs included one count each of pyrexia, duodenitis, increased transaminases and hyperthyroidism. The RP2D was 1012 viral particles (VP) ChAd68 and 30 µg samRNA. Secondary endpoints included immunogenicity, feasibility of manufacturing and overall survival (OS). Vaccine manufacturing was feasible, with vaccination inducing long-lasting neoantigen-specific CD8 T cell responses. Several patients with microsatellite-stable colorectal cancer (MSS-CRC) had improved OS. Exploratory biomarker analyses showed decreased circulating tumor DNA (ctDNA) in patients with prolonged OS. Although small study size limits statistical and translational analyses, the increased OS observed in MSS-CRC warrants further exploration in larger randomized studies.
(Methods section) cfDNA was extracted from the entire plasma volume of a single draw using the Apostle MiniMax cfDNA Isolation kit (ApostleBio)
Science Translational Medicine 2022
- Simultaneous analysis of mutations and methylations in circulating cell-free DNA for hepatocellular carcinoma detection. Pei Wan, Qianqian Son, Jie Ren, et al. Science Translational Medicine 14, eabp8704 (2022) 23 November 2022
(Note: Apostle MiniMax technology is used in this study.)
Cell-free DNA (cfDNA)–based liquid biopsy is a promising approach for the early detection of cancer. A major hurdle is the limited yield of cfDNA from one blood draw, limiting the use of most samples to one test of either mutation or methylation. Here, we develop a technology, Mutation Capsule Plus (MCP), which enables multiplex profiling of one cfDNA sample, including simultaneous detection of genetic and epigenetic alterations and genome-wide discovery of methylation markers. With this technology, we performed de novo screening of methylation markers on cfDNA samples from 30 hepatocellular carcinoma (HCC) cases and 30 non-HCC controls. The methylation markers enriched in HCC cfDNA were further profiled in parallel with a panel of mutations on a training cohort of 60 HCC and 60 non-HCC cases, resulting in an HCC detection model. We validated the model in an independent retrospective cohort with 58 HCC and 198 non-HCC cases and got 90% sensitivity with 94% specificity. Furthermore, we applied the model to a prospective cohort of 311 asymptomatic hepatitis B virus carriers with normal liver ultrasonography and serum AFP concentration. The model detected four of the five HCC cases in the cohort, showing 80% sensitivity and 94% specificity. These findings demonstrate that the MCP technology has potential for the discovery and validation of multiomics biomarkers for the noninvasive detection of cancer. This study also provides a comprehensive database of genetic and epigenetic alterations in the cfDNA of a large cohort of HCC cases and high-risk non-HCC individuals.
(Methods Section) cfDNA was extracted from the plasma samples using the Apostle MiniMax cfDNA isolation kit (C43468, Apostle).
Nature Medicine 2022
- Safety and tolerability of AAV8 delivery of a broadly neutralizing antibody in adults living with HIV: a phase 1, dose-escalation trial. Casazza JP, Cale EM, et al. the VRC603 Study Team. Nature Medicine April 11, 2022; https://www.nature.com/articles/s41591-022-01762-x
(Note: Apostle MiniMax technology is used in this study.)
Adeno-associated viral vector-mediated transfer of DNA coding for broadly neutralizing anti-HIV antibodies (bnAbs) offers an alternative to attempting to induce protection by vaccination or by repeated infusions of bnAbs. In this study, we administered a recombinant bicistronic adeno-associated virus (AAV8) vector coding for both the light and heavy chains of the potent broadly neutralizing HIV-1 antibody VRC07 (AAV8-VRC07) to eight adults living with HIV. All participants remained on effective anti-retroviral therapy (viral load (VL) <50 copies per milliliter) throughout this phase 1, dose-escalation clinical trial (NCT03374202). AAV8-VRC07 was given at doses of 5 × 1010, 5 × 1011 and 2.5 × 1012 vector genomes per kilogram by intramuscular (IM) injection. Primary endpoints of this study were to assess the safety and tolerability of AAV8-VRC07; to determine the pharmacokinetics and immunogenicity of in vivo VRC07 production; and to describe the immune response directed against AAV8-VRC07 vector and its products. Secondary endpoints were to assess the clinical effects of AAV8-VRC07 on CD4 T cell count and VL and to assess the persistence of VRC07 produced in participants. In this cohort, IM injection of AAV8-VRC07 was safe and well tolerated. No clinically significant change in CD4 T cell count or VL occurred during the 1–3 years of follow-up reported here. In participants who received AAV8-VRC07, concentrations of VRC07 were increased 6 weeks (P = 0.008) and 52 weeks (P = 0.016) after IM injection of the product. All eight individuals produced measurable amounts of serum VRC07, with maximal VRC07 concentrations >1 µg ml−1 in three individuals. In four individuals, VRC07 serum concentrations remained stable near maximal concentration for up to 3 years of follow-up. In exploratory analyses, neutralizing activity of in vivo produced VRC07 was similar to that of in vitro produced VRC07. Three of eight participants showed a non-idiotypic anti-drug antibody (ADA) response directed against the Fab portion of VRC07. This ADA response appeared to decrease the production of serum VRC07 in two of these three participants. These data represent a proof of concept that adeno-associated viral vectors can durably produce biologically active, difficult-to-induce bnAbs in vivo, which could add valuable new tools to the fight against infectious diseases.
(Methods Section) AAV8-VRC07 vector DNA quantitation. Plasma AAV8-VRC07 plasmid DNA was measured by extracting DNA from plasma, concentrating and then using a real-time PCR assay to measure a 103 base sequence spanning the junction of the IgG heavy chain sequence and F2A insert. DNA was extracted from serum using an Apostle MiniMax High Efficiency cfDNA Isolation Kit, following the manufacturer’s protocol with slight modification.
PNAS 2021
- Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy. Shengnan Jin, Dewen Zhu, Fanggui Shao, et al. PNAS February 2, 2021 118 (5) e2017421118; https://doi.org/10.1073/pnas.2017421118
(Note: Apostle MiniMax technology is used in this study.)
Multiplex assays, involving the simultaneous use of multiple circulating tumor DNA (ctDNA) markers, can improve the performance of liquid biopsies so that they are highly predictive of cancer recurrence. We have developed a single-tube methylation-specific quantitative PCR assay (mqMSP) that uses 10 different methylation markers and is capable of quantitative analysis of plasma samples with as little as 0.05% tumor DNA. In a cohort of 179 plasma samples from colorectal cancer (CRC) patients, adenoma patients, and healthy controls, the sensitivity and specificity of the mqMSP assay were 84.9% and 83.3%, respectively. In a head-to-head comparative study, the mqMSP assay also performed better for detecting early-stage (stage I and II) and premalignant polyps than a published SEPT9 assay. In an independent longitudinal cohort of 182 plasma samples (preoperative, postoperative, and follow-up) from 82 CRC patients, the mqMSP assay detected ctDNA in 73 (89.0%) of the preoperative plasma samples. Postoperative detection of ctDNA (within 2 wk of surgery) identified 11 of the 20 recurrence patients and was associated with poorer recurrence-free survival (hazard ratio, 4.20; P = 0.0005). With subsequent longitudinal monitoring, 14 patients (70%) had detectable ctDNA before recurrence, with a median lead time of 8.0 mo earlier than seen with radiologic imaging. The mqMSP assay is cost-effective and easily implementable for routine clinical monitoring of CRC recurrence, which can lead to better patient management after surgery.
Best-in-Class Performance in cfDNA Isolation: Side by Side Comparison
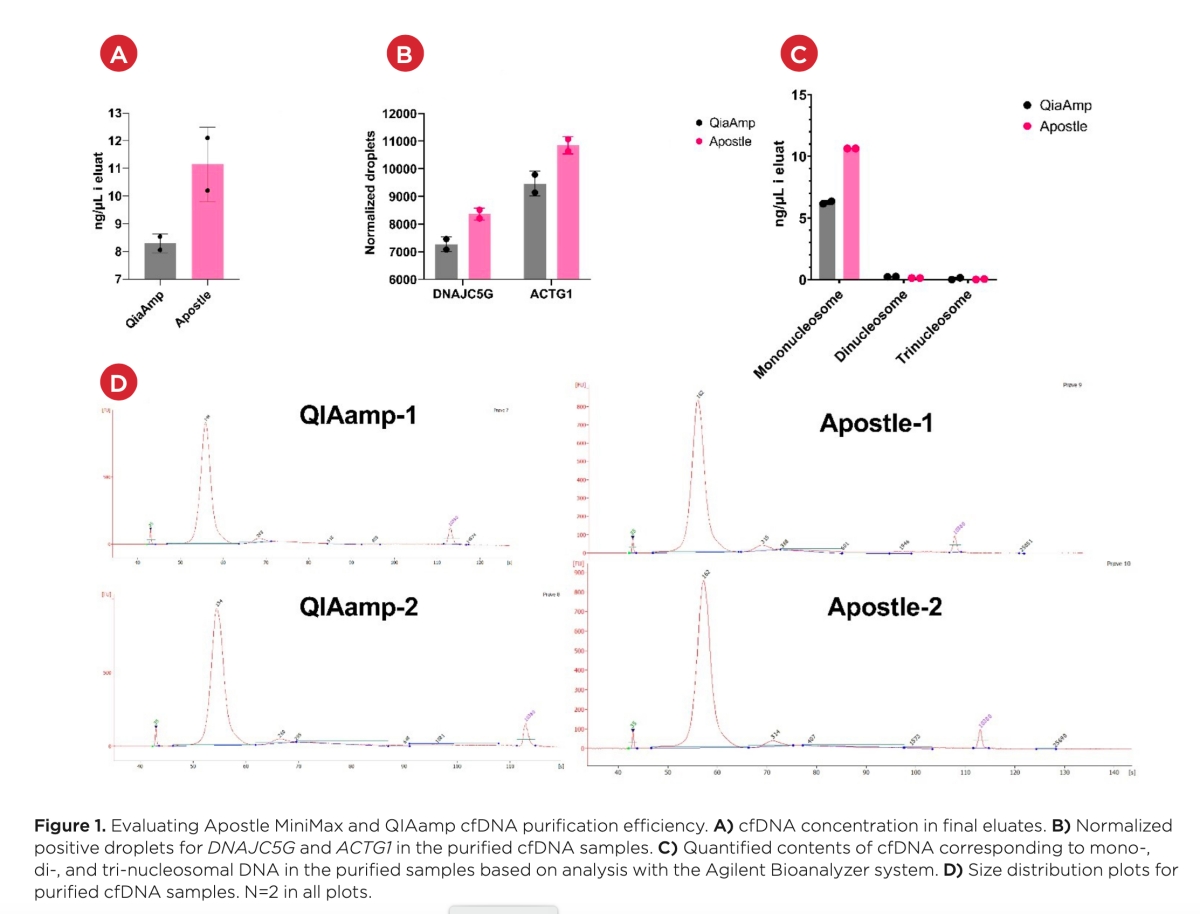
Results and discussion
Mean cfDNA concentrations in the eluates were determined to be 8.30 ng/μL for samples extracted with the QIAamp kit and 11.15 ng/μL for samples extracted with the Apostle kit when measured with the Qubit assay (Figure 1A). This illustrates a 34.3% higher yield when using the Apostle kit as compared to the QIAamp kit.
When evaluating the contents of specific genes in the eluate, the Apostle purification displayed a higher yield than QIAamp (Figure 1B). 8358 DNAJC5G positive droplets were detected in the Apostle samples compared to 7269 droplets in the QIAamp samples. Similarly, 10849 ACTG1 droplets were detected in the Apostle-extracted samples versus 9463 droplets detected in the QIAamp-extracted samples. This data demonstrates an average of 14.8% higher yield using the Apostle kit.
Finally, when evaluating cfDNA fragments via the Agilent bioanalyzer instrument, the Apostle kit demonstrated the highest yield (Figure 1C and D). Mononucleosomal cfDNA concentration in the eluate of Apostle samples was estimated to be 10.64 ng/μL versus 6.26 ng/μL in the eluate of QIAamp samples. The dinucleosomal cfDNA concentration was low for both kits, with 0.13 ng/μL recorded for Apostle samples and 0.24 ng/μL for QIAamp samples. This indicates a slightly better purification of dinucleosomal cfDNA using QIAamp; however, this could also be an artifact given the low concentrations. The Apostle mononucleosomal cfDNA yield was 70.0% higher than the yield for the QIAamp samples.
Publication: Comparative analysis of cell-free DNA extraction efficiency from plasma. Beckman Coulter Life Sciences. Indianapolis, IN. Technical Note 2022.
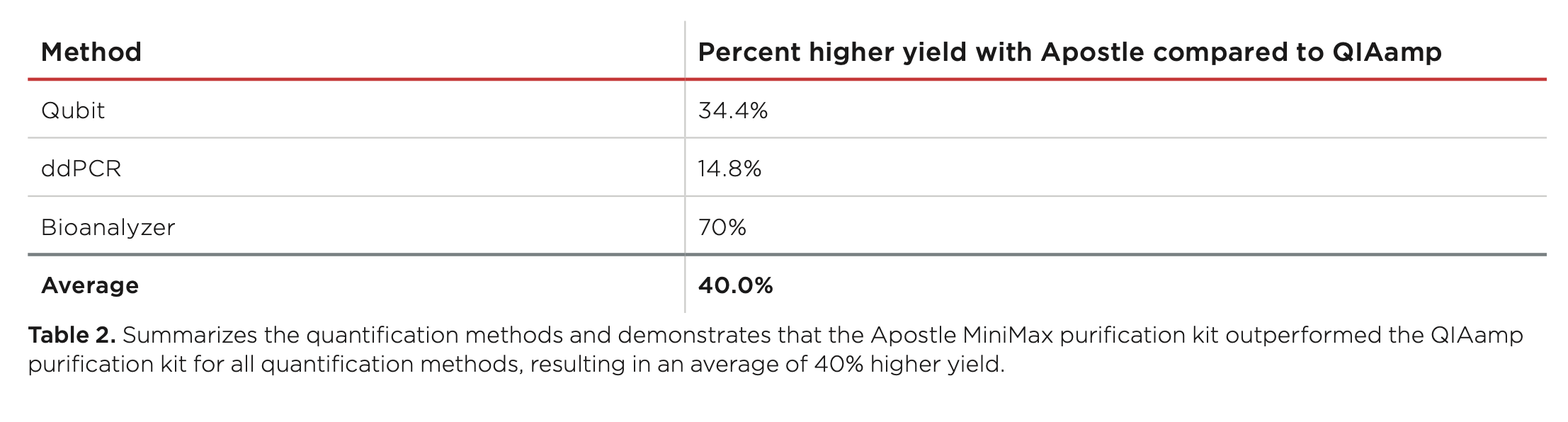
A summary provided in Table 2 shows the consistently higher yield achieved by the Apostle MiniMax High Efficiency cfDNA Isolation Kit as compared to the QIAamp® circulating nucleic acid kit. The average 40% improvement in yield underscores the importance of extraction kit selection in cfDNA assay development and highlights the potential impact of cfDNA isolation efficiency in ctDNA detection.
Publication: Comparative analysis of cell-free DNA extraction efficiency from plasma. Beckman Coulter Life Sciences. Indianapolis, IN. Technical Note 2022.
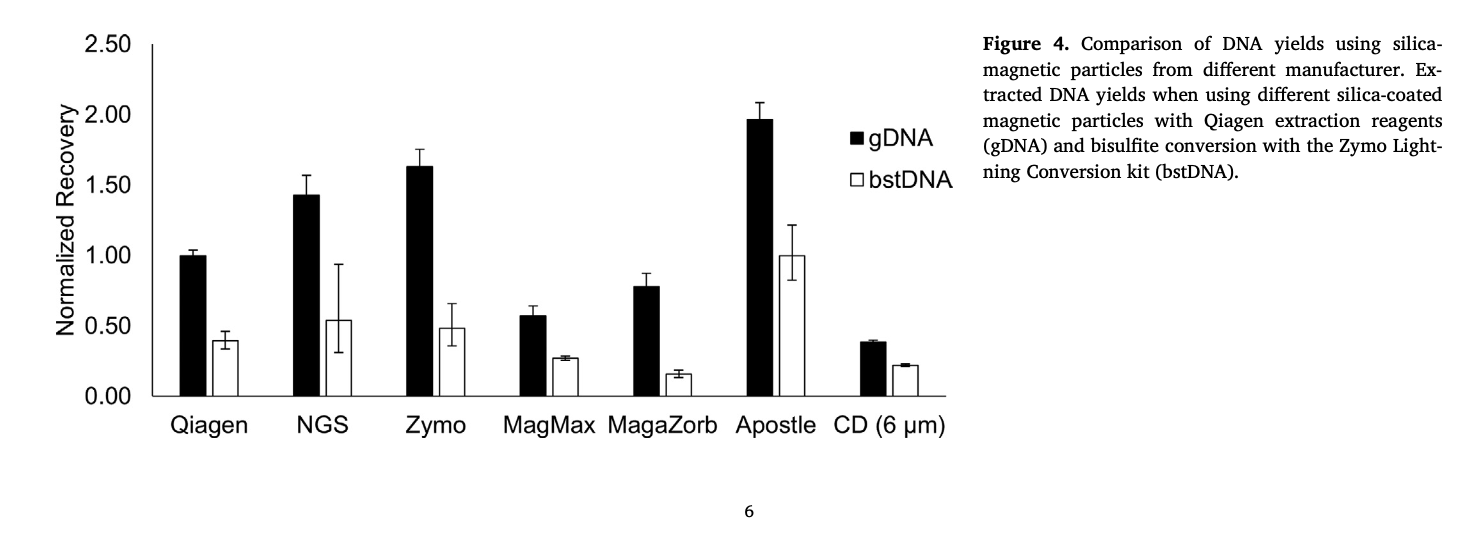
(Figure 4) - "Most notably, the Apostle particles outperformed all others, achieving almost 2-fold higher recovery yields than the particles supplied in the Qiagen kit. "
Publication: High-throughput sample processing for methylation analysis in an automated, enclosed environment. Stark et al. SLAS Technology 2022; 27(3):172-179.

Publication: Optimization of high-volume cell free DNA extraction and end-to-end automation for TSO 500 ctDNA library prep [abstract]. Nripesh Prasad, Rachel Rock, Rebecca Beatty, Melanie Robinson, Dineen Wildman, Michael Sykes, Boris Umylny, Thomas Halsey. In: Proceedings of the American Association for Cancer Research Annual Meeting 2024; Part 1 (Regular Abstracts); 2024 Apr 5-10; San Diego, CA. Philadelphia (PA): AACR; Cancer Res 2024;84(6_Suppl):Abstract nr 2298.
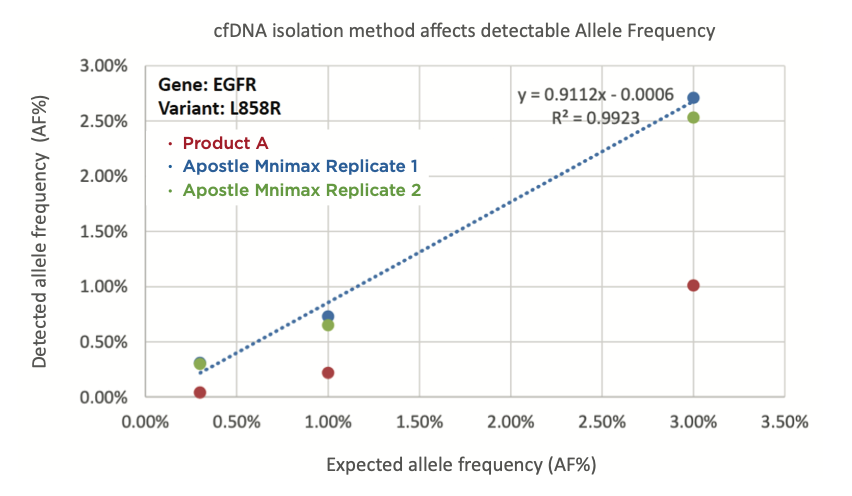
Figure 1. cfDNA extraction method affects allele frequency (AF%). EGFR L858R standards with AF% of 0.3%, 1.0%, and 3.0% were spiked into plasma and isolated using Product A (red) or Apostle MiniMax cfDNA isolation kit (blue and green). The eluates were analyzed by NGS (performed by the Institute B) following standard protocol and AF% calculated using their established workflow. cfDNA isolated by Apostle MiniMax cfDNA isolation kit shows higher concordance between detected AF% and expected AF%.
Publication: cfDNA Extraction Efficiency Affects NGS Data. Beckman Coulter Life Sciences. Indianapolis, IN. Technical Note 2020.
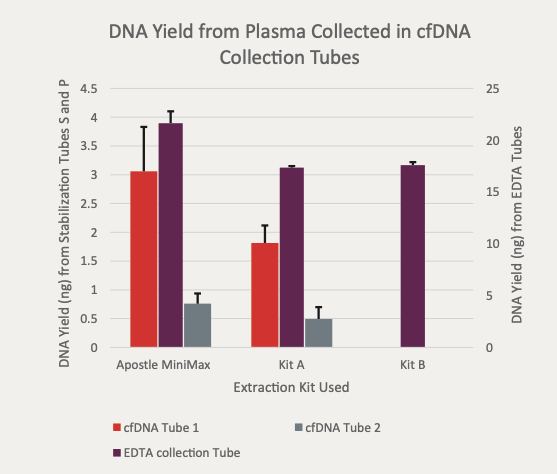
(Figure 1) "For each tube the Apostle MiniMax extracted higher total yield of DNA. "
Publication: cfDNA Extraction from Plasma for Liquid Biopsy: Apostle MiniMaxTM High Efficiency cfDNA Isolation Kit. Beckman Coulter Life Sciences, Data Sheet. 2019.
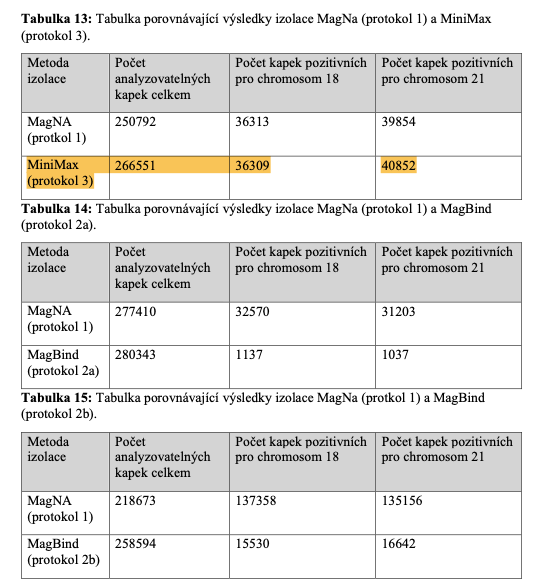
(Note: Table 13 referenced below in this diploma thesis shows that Apostle MiniMax yields the greatest number of analyzable drops in the digital PCR.)
Publication. Optimalizace digitální polymerázové řetězové reakce pro aplikaci v neinvazivní prenatální diagnostice (Optimization of digital polymerase chain reaction for application in non-invasive prenatal diagnostics) Author: Šenkyřík, Pavel; Advisor: Korabečná, Marie; Referee: Vodička, Radek; Faculty / Institute: Faculty of Science; Discipline: Anthropology and Human Genetics; Department: Department of Anthropology and Human Genetics; Date of defense: 13. 9. 2022; Publisher: Univerzita Karlova, Přírodovědecká fakulta; Language: Czech
Apostle technologies have been cited or discussed in the following publications.
Publications (2024)
67. A shared neoantigen vaccine combined with immune checkpoint blockade for advanced metastatic solid tumors: phase 1 trial interim results. Amy R. Rappaport, Chrisann Kyi, Monica Lane, et al. Nature Medicine March 27, 2024
(Note: Apostle MiniMax technology is used in this study.)
Abstract Therapeutic vaccines that elicit cytotoxic T cell responses targeting tumor-specific neoantigens hold promise for providing long-term clinical benefit to patients with cancer. Here we evaluated safety and tolerability of a therapeutic vaccine encoding 20 shared neoantigens derived from selected common oncogenic driver mutations as primary endpoints in an ongoing phase 1/2 study in patients with advanced/metastatic solid tumors. Secondary endpoints included immunogenicity, overall response rate, progression-free survival and overall survival. Eligible patients were selected if their tumors expressed one of the human leukocyte antigen-matched tumor mutations included in the vaccine, with the majority of patients (18/19) harboring a mutation in KRAS. The vaccine regimen, consisting of a chimp adenovirus (ChAd68) and self-amplifying mRNA (samRNA) in combination with the immune checkpoint inhibitors ipilimumab and nivolumab, was shown to be well tolerated, with observed treatment-related adverse events consistent with acute inflammation expected with viral vector-based vaccines and immune checkpoint blockade, the majority grade 1/2. Two patients experienced grade 3/4 serious treatment-related adverse events that were also dose-limiting toxicities. The overall response rate was 0%, and median progression-free survival and overall survival were 1.9 months and 7.9 months, respectively. T cell responses were biased toward human leukocyte antigen-matched TP53 neoantigens encoded in the vaccine relative to KRAS neoantigens expressed by the patients’ tumors, indicating a previously unknown hierarchy of neoantigen immunodominance that may impact the therapeutic efficacy of multiepitope shared neoantigen vaccines. These data led to the development of an optimized vaccine exclusively targeting KRAS-derived neoantigens that is being evaluated in a subset of patients in phase 2 of the clinical study. ClinicalTrials.gov registration: NCT03953235.
(Methods section)
cfDNA was extracted from the entire plasma volume of a single draw using the Apostle MiniMax cfDNA Isolation kit (ApostleBio) and quantified using the Qubit 1× dsDNA High Sensitivity Assay (Thermo Fisher Scientific).
66. Tumor- and circulating-free DNA methylation identifies clinically relevant small cell lung cancer subtypes. Simon Heeke, Carl M. Gay, Marcos R. Estecio, et al. Cancer Cell January 25, 2024
(Note: Apostle MiniMax technology is used in this study.)
Abstract Small cell lung cancer (SCLC) is an aggressive malignancy composed of distinct transcriptional subtypes, but implementing subtyping in the clinic has remained challenging, particularly due to limited tissue availability. Given the known epigenetic regulation of critical SCLC transcriptional programs, we hypothesized that subtype-specific patterns of DNA methylation could be detected in tumor or blood from SCLC patients. Using genomic-wide reduced-representation bisulfite sequencing (RRBS) in two cohorts totaling 179 SCLC patients and using machine learning approaches, we report a highly accurate DNA methylation-based classifier (SCLC-DMC) that can distinguish SCLC subtypes. We further adjust the classifier for circulating-free DNA (cfDNA) to subtype SCLC from plasma. Using the cfDNA classifier (cfDMC), we demonstrate that SCLC phenotypes can evolve during disease progression, highlighting the need for longitudinal tracking of SCLC during clinical treatment. These data establish that tumor and cfDNA methylation can be used to identify SCLC subtypes and might guide precision SCLC therapy.
(Methods section)
Critical commercial assays
Apostle MiniMax High Efficiency Cell-Free DNA Isolation Kit Apostle Bio A17622-250
Nucleic acid extraction
cfDNA was extracted using the Apostle MiniMax High Efficiency Cell-Free DNA Isolation Kit (Apostle Inc).
65. Terminal modifications independent cell-free RNA sequencing enables sensitive early cancer detection and classification. Jun Wang, Jinyong Huang, Yunlong Hu, et al. Nature Communications 15, Article number: 156 (2024)
(Note: Apostle MiniMax technology is used in this study.)
Abstract Cell-free RNAs (cfRNAs) offer an opportunity to detect diseases from a transcriptomic perspective, however, existing techniques have fallen short in generating a comprehensive cell-free transcriptome profile. We develop a sensitive library preparation method that is robust down to 100 µl input plasma to analyze cfRNAs independent of their 5’-end modifications. We show that it outperforms adapter ligation-based method in detecting a greater number of cfRNA species. We perform transcriptome-wide characterizations in 165 lung cancer, 30 breast cancer, 37 colorectal cancer, 55 gastric cancer, 15 liver cancer, and 133 cancer-free participants and demonstrate its ability to identify transcriptomic changes occurring in early-stage tumors. We also leverage machine learning analyses on the differentially expressed cfRNA signatures and reveal their robust performance in cancer detection and classification. Our work sets the stage for in-depth study of the cfRNA repertoire and highlights the value of cfRNAs as cancer biomarkers in clinical applications.
(Methods section) cfRNA extraction
Frozen plasma samples were thawed on ice prior to cfRNA extraction. 200 μl of plasma samples were subjected to cfRNA extraction using the Apostle MiniMax High-Efficiency cfRNA Isolation Kit (Apostle), following the manufacturer’s protocol with minor modifications.
64. A Blood Hepatocellular Carcinoma Signature Recognizes Very Small Tumor Nodules with Metastatic Traits. Kun Chen, Junxiao Wang, Liping Jiang, et al. Journal of Clinical and Translational Hepatology 2024, doi: 10.14218/JCTH.2023.00559
(Note: Apostle MiniMax technology is used in this study.)
Abstract
Background and Aims
Hepatocellular carcinoma (HCC) cases with small nodules are commonly treated with radiofrequency ablation (RFA), but the recurrence rate remains high. This study aimed to establish a blood signature for identifying HCC with metastatic traits pre-RFA.
Methods
Data from HCC patients treated between 2010 and 2017 were retrospectively collected. A blood signature for metastatic HCC was established based on blood levels of alpha-fetoprotein and des-γ-carboxy-prothrombin, cell-free DNA (cfDNA) mutations, and methylation changes in target genes in frozen-stored plasma samples that were collected before RFA performance. The HCC blood signature was validated in patients prospectively enrolled in 2021.
Results
Of 251 HCC patients in the retrospective study, 33.9% experienced recurrence within 1 year post-RFA. The HCC blood signature identified from these patients included des-γ-carboxy-prothrombin ≥40 mAU/mL with cfDNA mutation score, where cfDNA mutations occurred in the genes of TP53, CTNNB1, and TERT promoter. This signature effectively predicted 1-year post-RFA recurrence of HCC with 92% specificity and 91% sensitivity in the retrospective dataset, and with 87% specificity and 76% sensitivity in the prospective dataset (n=32 patients). Among 14 cases in the prospective study with biopsy tissues available, positivity for the HCC blood signature was associated with a higher HCC tissue score and shorter distance between HCC cells and microvasculature.
Conclusions
This study established an HCC blood signature in pre-RFA blood that potentially reflects HCC with metastatic traits and may be valuable for predicting the disease’s early recurrence post-RFA.
(Methods section)
Analysis of cfDNA mutations and methylation changes in stored plasma
Briefly, cfDNA was extracted from the plasma samples using the Apostle MiniMax cfDNA isolation kit. The cfDNA (5–40 ng) was digested using the methylation-sensitive restriction enzyme Hha I (R0139L, New England BioLabs) and subsequently ligated to customized MCP adapters with random DNA barcodes.
63. Optimization of high-volume cell free DNA extraction and end-to-end automation for TSO 500 ctDNA library prep [abstract]. Nripesh Prasad, Rachel Rock, Rebecca Beatty, Melanie Robinson, Dineen Wildman, Michael Sykes, Boris Umylny, Thomas Halsey. In: Proceedings of the American Association for Cancer Research Annual Meeting 2024; Part 1 (Regular Abstracts); 2024 Apr 5-10; San Diego, CA. Philadelphia (PA): AACR; Cancer Res 2024;84(6_Suppl):Abstract nr 2298.
(Note: Apostle MiniMax technology is used and compared in this study.)
Abstract Liquid biopsy such as plasma, has emerged as one of the most valuable sample types for profiling circulating tumor DNA (ctDNA) for screening, response to treatment, and monitoring relapse in oncology patients. Due to the diffuse nature of ctDNA however, both extraction and library preparation must be optimized for the efficient downstream next-generation sequencing (NGS) application. We initially addressed the optimization of quality and quantity of cfDNA recovery from healthy and various oncology patients by evaluating QIAsymphony DSP Circulating DNA Kit and Apostle MiniMax cfDNA isolation kits. Overall, the cfDNA yields from Apostle MiniMax kit were approximately 12X higher. We then optimized high-volume automated cfDNA extraction from 1ml, 3ml and 6ml plasma samples using the Biomek i7 for Apostle MiniMax kit. Colorectal, Gastric and Ovarian cancer gave the highest yield of cfDNA whereas, Esophageal, Lung and Kidney cancers were the lowest cfDNA yielding samples. We then analyzed ctDNA from plasma from healthy donors, colorectal, kidney, Breast, and endometrial cancer patients along with Oncospan and Seraseq ctDNA mutation mix (5-0.5% AF) to evaluate the mutation detection efficacy, repeatability, and reproducibility by targeted NGS using TSO 500ctDNA assay ay 10ng, 20ng, and 30ng inputs. For library preparation batches of 48 samples were prepared using a fully automated processes for the library prep, Hybridization capture and post capture enrichment on the Beckman i5 robot. Libraries were sequenced on NovaSeq 6000 at 600M Paired-end depth. This method demonstrates an overall concordance of inter-run and intra-run accuracy of variant detection to be 90% demonstrating that this method run as described produces highly repeatable and reproducible variant detection to sub-1% allele frequencies. The inter-run, lot-to-lot, and operator-to-operator variability was ≥99%. Down-sample analysis demonstrated high levels of assay performance with sample inputs of 30 ng with ≥96% sample passing Illumina cut-off metrics at 150M PE read depth as compared to Illumina recommended 420M PE reads. Taken together, we provide a high-volume automated solution for cfDNA extraction from plasma samples, additionally these data demonstrate a robust, reproducible, and highly accurate targeted DNA sequencing method that can be used for low allele frequency variant detection in cfDNA.
62. Sequencing of cerebrospinal fluid cell-free DNA facilitated early differential diagnosis of intramedullary spinal cord tumors. Chai, et al. npj Precision Oncology volume 8, Article number: 43 (2024)
(Note: Apostle MiniMax technology is used in this study.)
Abstract
Pre-surgery differential diagnosis is valuable for personalized treatment planning in intramedullary spinal cord tumors. This study assessed the performance of sequencing cell-free DNA (cfDNA) in cerebrospinal fluid (CSF) for differential diagnosis of these tumors. Prospectively enrolling 45 patients with intramedullary spinal cord lesions, including diffuse midline glioma (DMG), H3K27-altered (14/45), glioblastoma (1/45), H3-wildtype-astrocytoma (10/45), ependymoma (11/45), and other lesions (9/45), CSF samples were collected via lumbar puncture (41/45), intraoperative extraction (3/45), and Ommaya reservoir (1/45). Then, these samples underwent targeted sequencing along with paired tissue DNA. DMG, H3K27-altered patients exhibited a higher ctDNA positivity (85.7%, 12/14) compared to patients with H3-wildtype-astrocytoma (0/8, P = 0.0003), ependymoma (2/10, P = 0.003), and glioneuronal tumor (0/3, P = 0.009). The histological-grade-IV (P = 0.0027), Ki-67 index ≥10% (P = 0.014), and tumor reaching spinal cord surface (P = 0.012) are also associated with higher ctDNA positivity. Interestingly, for patients with TERT promoter mutant tumors, TERT mutation was detectable in the CSF cfDNA of one DMG case, but not other five cases with histological-grade-II tumors. Shared copy number variants were exclusively observed in DMG, H3K27-altered, and showed a strong correlation (Correlation = 0.95) between CSF and tissue. Finally, H3K27M mutations in CSF exhibited high diagnostic efficiency for DMG, H3K27-altered (Sensitivity = 85.7%, Specificity = 100.0%, AUC = 0.929). Notably, H3K27M was detectable in CSF from patients with recurrent tumors, making it easily applicable for postoperative monitoring. In conclusion, the molecular profile from ctDNA released into CSF of malignant tumors was more frequently detected compared to relatively benign ones. Sequencing of ctDNA in CSF exhibited high efficiency for the differential diagnosis of DMG, H3K27-altered.
(Methods section) Circulating cell-free DNA (cfDNA) isolation from cerebrospinal fluid (CSF)
Then, circulating nucleic acid was extracted from CSF using the Apostle MiniMax High-Efficiency cfDNA Isolation Kit (Apostle, USA) following the manufacturer's instructions.
61. A noninvasive multianalytical approach establishment for risk assessment and gastric cancer screening. Fan, et al. International Journal of Cancer, 2024; 154(6):1111-1123
(Note: Apostle MiniMax technology is used in this study.)
Abstract
Effective screening and early detection are critical to improve the prognosis of gastric cancer (GC). Our study aims to explore noninvasive multianalytical biomarkers and construct integrative models for preliminary risk assessment and GC detection. Whole genomewide methylation marker discovery was conducted with CpG tandems target amplification (CTTA) in cfDNA from large asymptomatic screening participants in a high-risk area of GC. The methylation and mutation candidates were validated simultaneously using one plasma from patients at various gastric lesion stages by multiplex profiling with Mutation Capsule Plus (MCP). Helicobacter pylori specific antibodies were detected with a recomLine assay. Integrated models were constructed and validated by the combination of multianalytical biomarkers. A total of 146 and 120 novel methylation markers were found in CpG islands and promoter regions across the genome with CTTA. The methylation markers together with the candidate mutations were validated with MCP and used to establish a 133-methylation-marker panel for risk assessment of suspicious precancerous lesions and GC cases and a 49-methylation-marker panel as well as a 144-amplicon-mutation panel for GC detection. An integrated model comprising both methylation and specific antibody panels performed better for risk assessment than a traditional model (AUC, 0.83 and 0.63, P < .001). A second model for GC detection integrating methylation and mutation panels also outperformed the traditional model (AUC, 0.82 and 0.68, P = .005). Our study established methylation, mutation and H. pylori-specific antibody panels and constructed two integrated models for risk assessment and GC screening. Our findings provide new insights for a more precise GC screening strategy in the future.
(Methods section) Blood sample collection and DNA extraction
The Apostle MiniMax cfDNA isolation kit (Apostle; San Jose, CA) was used for cfDNA extraction.
60. Dissecting patterns of small cell lung cancer evolution using deep whole genome sequencing of circulating tumor DNA [abstract]. Benjamin B. Morris, Zhihui Zhang, Simon Heeke, et al. In: Proceedings of the AACR Special Conference in Cancer Research: Translating Cancer Evolution and Data Science: The Next Frontier; 2023 Dec 3-6; Boston, Massachusetts. Philadelphia (PA): AACR; Cancer Res 2024;84(3 Suppl_2):Abstract nr A022.
(Note: Apostle MiniMax technology is used in this study.)
Abstract
Background: Small cell lung cancer (SCLC) is the most lethal form of lung cancer. A key driver of this near universal lethality is SCLC’s recalcitrance to therapy. The standard of care for SCLC is a chemotherapy doublet of etoposide and cis/carboplatin in combination with immunotherapy (EP+IO). While most tumors respond to these therapies, >80% of SCLCs progress within one year of treatment. Additionally, the aggressive clinical course of SCLC has historically precluded collection of patient-matched treatment naïve and recurrent tumor tissue traditionally required to study evolution. In lieu of tissue, several groups have used circulating tumor DNA (ctDNA) to study SCLC evolution. However, these studies used targeted sequencing panels that analyze <0.04% of the SCLC genome. Using new methods to comprehensively trace how SCLCs rapidly evolve to therapy resistance is a significant unmet need.
Methods: Medical records were reviewed to identify treatment naïve SCLC patients treated at MD Anderson that had both pre- and post-EP+IO liquid biopsies collected under IRB approved protocols. ctDNA was isolated from plasma using a MiniMax High Efficiency Cell-Free DNA Isolation Kit (Apostle Bio). Germline DNA was extracted from buffy coat samples using a QIAamp DNA Blood Midi Kit (Qiagen). Deep whole genome sequencing (WGS) (>100X) of ctDNA and WGS of germline DNA (>60X) was performed using an Illumina NovaSeq X sequencer. Reads were aligned to the hg38 reference genome using BWA-mem. Somatic mutations were called using MuTect2, and clonal and subclonal copy number alterations were identified using Battenberg. DPClust was used to reconstruct cancer cell populations, leveraging mutation and copy number calls.
Results: We identified several deleterious genomic alterations impacting TP53 and RB1 tumor suppressor genes, known genetic drivers of SCLC development. We also detected recurrent, whole chromosome arm loss of heterozygosity events impacting many chromosomes in treatment naïve samples, some of which are known to be present in >80% of SCLCs. Additionally, we found that both treatment naïve and recurrent SCLCs were composed of multiple subclones, each defined by dozens to thousands of unique somatic mutations. COSMIC signature analysis identified high activity of tobacco carcinogens, APOBEC, and replicative-based processes across ctDNA samples. We also identified emergence of novel subclones carrying unique copy number alterations following EP+IO recurrence. Importantly, the affected regions encode key effectors that regulate anti-tumor immune responses and cell survival following DNA damage, both of which are intimately related to the mechanism of action of EP+IO.
Conclusions: Our data shows that deep WGS of ctDNA recapitulates known genetics of SCLC and allows for sensitive characterization of therapy resistant clones. Using this method, we have captured snapshots of both intrinsic and acquired resistance mechanisms to frontline EP+IO. Deep WGS of ctDNA is a promising approach to comprehensively dissect routes of SCLC evolution and therapy resistance.
59. Comprehensive characterization of small noncoding RNA profiles in hypoxia-induced pulmonary hypertension (HPH) rat tissues. Jun Wang, Jiahao Kuang, Shasha Zhang, et al. iScience 2024 Feb 16; 27(2): 108815.
(Note: Apostle MiniMax technology is used in this study.)
Abstract
Hypoxia-induced pulmonary hypertension (HPH) is a fatal cardiovascular disease characterized by an elevation in pulmonary artery pressure, resulting in right ventricular dysfunction and eventual heart failure. Exploring the pathogenesis of HPH is crucial, and small noncoding RNAs (sncRNAs) are gaining recognition as potential regulators of cellular responses to hypoxia. In this study, we conducted a comprehensive analysis of sncRNA profiles in eight tissues of male HPH rats using high-throughput sequencing. Our study unveiled several sncRNAs, with the brain, kidney, and spleen exhibiting the highest abundance of microRNA (miRNA), tRNA-derived small RNA (tDR), and small nucleolar RNA (snoRNA), respectively. Moreover, we identified numerous tissue-specific and hypoxia-responsive sncRNAs, particularly miRNAs and tDRs. Interestingly, we observed arm switching in miRNAs under hypoxic conditions and a significant increase in the abundance of 5′ tRNA-halves among the total tDRs during hypoxia. Overall, our study provides a comprehensive characterization of the sncRNA profiles in HPH rats.
Methods Section
sncRNA sequencing library preparation
To isolate total RNA from the eight different tissues and plasma, we used RNAiso Plus (Takara, Japan) and Apostle MiniMax High Efficiency cfRNA Isolation Kit (Apostle) according to the manufacturer’s instructions.
58. Ultra-sensitive molecular residual disease detection through whole genome sequencing with single-read error correction. Xinxing Li, Tao Liu, Antonella Bacchiocchi, et al. medRxiv January 2024, preprint doi: https://doi.org/10.1101/2024.01.13.24301070
(Note: Apostle MiniMax technology is used in this study.)
Abstract While whole genome sequencing (WGS) of cell-free DNA (cfDNA) holds enormous promise for molecular residual disease (MRD) detection, its performance is limited by WGS error rate. Here we introduce AccuScan, an efficient cfDNA WGS technology that enables genome-wide error correction at single read level, achieving an error rate of 4.2×10-7, which is about two orders of magnitude lower than a read-centric de-noising method. When applied to MRD detection, AccuScan demonstrated analytical sensitivity down to 10-6 circulating tumor allele fraction at 99% sample level specificity. In colorectal cancer, AccuScan showed 90% landmark sensitivity for predicting relapse. It also showed robust MRD performance with esophageal cancer using samples collected as early as 1 week after surgery, and predictive value for immunotherapy monitoring with melanoma patients. Overall, AccuScan provides a highly accurate WGS solution for MRD, empowering circulating tumor DNA detection at parts per million range without high sample input nor personalized reagents.
(Methods section) Plasma DNA processing
Isolation of cfDNA from 1 ~ 2 mL of plasma was performed using MiniMax High Efficiency Cell-Free Isolation Kit (catalog no. A17622CN-384, Apostle) and eluted in 80 μL Tris-EDTA buffer.
57. Integration of Cell-Free DNA End Motifs and Fragment Lengths Can Identify Active Genes in Liquid Biopsies. Christoffer Trier Maansson, Louise Skov Thomsen, Peter Meldgaard, et al. International Journal of Molecular Sciences. 2024, 25(2), 1243
(Note: Apostle MiniMax technology is used in this study.)
Abstract Multiple studies have shown that cell-free DNA (cfDNA) from cancer patients differ in both fragment length and fragment end motif (FEM) from healthy individuals, yet there is a lack of understanding of how the two factors combined are associated with cancer and gene transcription. In this study, we conducted cfDNA fragmentomics evaluations using plasma from lung cancer patients (n = 12) and healthy individuals (n = 7). A personal gene expression profile was established from plasma using H3K36me3 cell-free chromatin immunoprecipitation sequencing (cfChIP-seq). The genes with the highest expression displayed an enrichment of short cfDNA fragments (median = 19.99%, IQR: 16.94–27.13%, p < 0.0001) compared to the genes with low expression. Furthermore, distinct GC-rich FEMs were enriched after cfChIP. Combining the frequency of short cfDNA fragments with the presence of distinct FEMs resulted in an even further enrichment of the most expressed genes (median = 37.85%, IQR: 30.10–39.49%, p < 0.0001). An in vitro size selection of <150 bp cfDNA could isolate cfDNA representing active genes and the size-selection enrichment correlated with the cfChIP-seq enrichment (Spearman r range: 0.499–0.882, p < 0.0001). This study expands the knowledge regarding cfDNA fragmentomics and sheds new light on how gene activity is associated with both cfDNA fragment lengths and distinct FEMs.
(Methods section) Cell-Free Chromatin Immunoprecipitaiton (cfChIP) Enrichment
The cfChIP sample and the input plasma cfDNA were purified using an Apostle MiniMax High Efficiency cfDNA Isolation Kit (Beckman Coulter, Indianapolis, IN, USA) and subjected to CAPP-seq.
Publications (2023)
56. Integrative modeling of tumor genomes and epigenomes for enhanced cancer diagnosis by cell-free DNA. Mingyun Bae, Gyuhee Kim, Tae-Rim Lee, et al. Nature Communications 14, Article number: 2017 (2023)
(Note: Apostle MiniMax technology is used in this study.)
Abstract Multi-cancer early detection remains a key challenge in cell-free DNA (cfDNA)-based liquid biopsy. Here, we perform cfDNA whole-genome sequencing to generate two test datasets covering 2125 patient samples of 9 cancer types and 1241 normal control samples, and also a reference dataset for background variant filtering based on 20,529 low-depth healthy samples. An external cfDNA dataset consisting of 208 cancer and 214 normal control samples is used for additional evaluation. Accuracy for cancer detection and tissue-of-origin localization is achieved using our algorithm, which incorporates cancer type-specific profiles of mutation distribution and chromatin organization in tumor tissues as model references. Our integrative model detects early-stage cancers, including those of pancreatic origin, with high sensitivity that is comparable to that of late-stage detection. Model interpretation reveals the contribution of cancer type-specific genomic and epigenomic features. Our methodologies may lay the groundwork for accurate cfDNA-based cancer diagnosis, especially at early stages.
(Methods section) cfDNA was extracted from 0.4 mL plasma ... and eluted in a final volume of 22 μL, using an Apostle MiniMax High Efficiency cfDNA Isolation Kit (Apostle, US) according to the manufacturer’s instructions.
55. Comprehensive Analysis of Cell-Free DNA Fragmentation Across Cancer Stages. Guo, et al. medRxiv, November 2023; https://doi.org/10.1101/2023.11.07.23298181
(Note: Apostle MiniMax technology is used in this study.)
Abstract
Circulating cell-free DNA (cfDNA) in the bloodstream displays cancer-derived fragmentation patterns, offering a non-invasive diagnostic avenue for cancer patients. However, the association between cfDNA fragmentation patterns and cancer progression remains largely unexplored. In this study, we analyzed this relationship using 214 whole-genome cfDNA samples across seven solid cancer types and revealed that the relation between cfDNA fragmentation patterns and cancer stages vary across cancer types. Among them, cfDNA fragmentation patterns in colorectal cancer (CRC) showed a strong correlation with cancer stages. This finding is further validated with an independent targeted cfDNA dataset from 29 CRC samples. Inspired by these findings, we designed “frag2stage”, a machine learning model that exploits cfDNA fragmentation data to differentiate cancer stages of CRC. Evaluated on two independent cfDNA datasets, our model can distinguish cancer stages of CRC with the area under the curve (AUC) values ranging from 0.68 to 0.99. The results suggest that cfDNA fragmentation patterns might carry yet undiscovered genetic and epigenetic signals, highlighting their promising potential for broader diagnostic applications in oncology.
(Methods section) CFDNA EXTRACTION AND NGS LIBRARY PREPARATION
Blood samples, about 10 ml from each individual, were collected using the Apostle MiniMax cf-DNA Blood Collection Tube (Apostle; San Jose, CA, USA) and processed within seven days from the collection. The cfDNA extraction from 2 to 5 ml of each plasma sample was conducted using the Apostle MiniMax High Efficiency cfDNA isolation kit (Apostle; San Jose, CA, USA), with adherence to the manufacturer’s protocol with slight modifications.
54. Concordance analysis of cerebrospinal fluid with the tumor tissue for integrated diagnosis in gliomas based on next-generation sequencing. Wang, et al. Pathology and Oncology Research, 26 September 2023 https://doi.org/10.3389/pore.2023.1611391
(Note: Apostle MiniMax technology is used in this study.)
Abstract
Purpose: The driver mutations of gliomas have been identified in cerebrospinal fluid (CSF). Here we compared the concordance between CSF and tumor tissue for integrated diagnosis in gliomas using next-generation sequencing (NGS) to evaluate the feasibility of CSF detection in gliomas.
Patients and methods: 27 paired CSF/tumor tissues of glioma patients were sequenced by a customized gene panel based on NGS. All CSF samples were collected through lumbar puncture before surgery. Integrated diagnosis was made by analysis of histology and tumor DNA molecular pathology according to the 2021 WHO classification of the central nervous system tumors.
Results: A total of 24 patients had detectable circulating tumor DNA (ctDNA) and 22 had at least one somatic mutation or chromosome alteration in CSF. The ctDNA levels varied significantly across different ages, Ki-67 index, magnetic resonance imaging signal and glioma subtypes (p < 0.05). The concordance between integrated ctDNA diagnosis and the final diagnosis came up to 91.6% (Kappa, 0.800). We reclassified the clinical diagnosis of 3 patients based on the results of CSF ctDNA sequencing, and 4 patients were reassessed depending on tumor DNA. Interestingly, a rare IDH1 R132C was identified in CSF ctDNA, but not in the corresponding tumor sample.
Conclusion: This study demonstrates a high concordance between integrated ctDNA diagnosis and the final diagnosis of gliomas, highlighting the practicability of NGS based detection of mutations of CSF in assisting integrated diagnosis of gliomas, especially glioblastoma.
(Methods section) DNA extraction and quantification
Cell-free DNA (cfDNA) was extracted using an Apostle MiniMax High Efficiency cfDNA Isolation Kit (APOSTLE) according to the manufacturer’s instructions.
53. Altered cfDNA fragmentation profile in hypomethylated regions as diagnostic markers in breast cancer. Wang J, et al. Epigenetics & Chromatin. 16, Article number: 33 (2023)
(Note: Apostle MiniMax technology is used in this study.)
Abstract
Background
Breast cancer, the most common malignancy in women worldwide, has been proven to have both altered plasma cell-free DNA (cfDNA) methylation and fragmentation profiles. Nevertheless, simultaneously detecting both of them for breast cancer diagnosis has never been reported. Moreover, although fragmentation pattern of cfDNA is determined by nuclease digestion of chromatin, structure of which may be affected by DNA methylation, whether cfDNA methylation and fragmentation are biologically related or not still remains unclear.
Methods
Improved cfMeDIP-seq were utilized to characterize both cfDNA methylation and fragmentation profiles in 49 plasma samples from both healthy individuals and patients with breast cancer. The feasibility of using cfDNA fragmentation profile in hypo- and hypermethylated regions as diagnostic markers for breast cancer was evaluated.
Results
Mean size of cfDNA fragments (100–220 bp) mapped to hypomethylated regions decreased more in patients with breast cancer (4.60 bp, 172.33 to 167.73 bp) than in healthy individuals (2.87 bp, 174.54 to 171.67 bp). Furthermore, proportion of short cfDNA fragments (100–150 bp) in hypomethylated regions when compared with it in hypermethylated regions was found to increase more in patients with breast cancer in two independent discovery cohort. The feasibility of using abnormality of short cfDNA fragments ratio in hypomethylated genomic regions for breast cancer diagnosis in validation cohort was evaluated. 7 out of 11 patients were detected as having breast cancer (63.6% sensitivity), whereas no healthy individuals were mis-detected (100% specificity).
Conclusion
We identified enriched short cfDNA fragments after 5mC-immunoprecipitation (IP) in patients with breast cancer, and demonstrated the enriched short cfDNA fragments might originated from hypomethylated genomic regions. Furthermore, we proved the feasibility of using differentially methylated regions (DMRs)-dependent cfDNA fragmentation profile for breast cancer diagnosis.
(Methods section) Sample collection and cfDNA extraction
cfDNA was extracted from plasma using MiniMax(TM) High Efficiency Cell-Free DNA Isolation Kit (Apostle, A17622-250) according to manufacturer’s instructions.
52. Combined detection of SDC2/ADHFE1/PPP2R5C methylation in stool DNA for colorectal cancer screening. Li B, et al. Journal of Cancer Research and Clinical Oncology. June 03, 2023. https://doi.org/10.1007/s00432-023-04943-4
(Note: Apostle MiniGenomics technology is used in this study.)
Abstract
Background Colorectal cancer (CRC) is a disease of global concern, and its increasing incidence suggests the need for early and accurate diagnosis. The aim of this study was to investigate the value of combined detection of SDC2, ADHFE1 and PPP2R5C gene methylation in stool samples for early CRC screening.
Methods StoolsamplesfrompatientswithCRC(n=105),advancedadenoma(AA)(n=54),non-advancedadenoma(NA) (n = 57), hyperplastic or other polyps (HOP) (n = 47) or no evidence of disease (NED) (n = 100) were collected from Sep- tember 2021 to September 2022. The methylation levels of SDC2, ADHFE1 and PPP2R5C were quantified by quantitative methylation-specific polymerase chain reaction (qMSP), and faecal immunochemical testing (FIT) was performed. The diagnostic value was assessed using reporter operating characteristic (ROC) curve analysis.
Results The sensitivity of combined detection of SDC2/ADHFE1/PPP2R5C methylation in predicting CRC (0–IV) was 84.8%, the specificity was 98.0%, and the AUC was 0.930 (95% CI 0.889–0.970). Compared to FIT and serum tumour bio- markers, it showed better diagnostic performance for different stages of CRC.
Conclusion The results of this study verified that the methylation levels of SDC2, ADHFE1 and PPP2R5C in stool DNA were significantly increased in CRC patients. Combined detection of SDC2/ADHFE1/PPP2R5C methylation is a potential non-invasive diagnostic method for CRC and precancerous lesion screening.
Clinical trial registration Chinese Clinical Trials Registry, ChiCTR2100046662, registered on 26 May 2021, prospective registration.
(Methods section) DNA isolation, bisulphite treatment and fluorescence‐based quantitative methylation‐specific polymerase chain reaction (qMSP)
Genomic DNA was isolated from stool samples using the Apostle MiniGenomics (TM) Stool Fast Kit (Apostle, A181206) and stored in the refrigerator at – 20 °C until further use.
51. Combining cell-free RNA with cell-free DNA in liquid biopsy for hematologic and solid tumors. Maher Albitar, Hong Zhang, Ahmad Charifa, et al. Heliyon 9 (2023) e16261; May 16, 2023
(Note: Apostle MiniMax technology is used in this study.)
Abstract Current use of liquid biopsy is based on cell-free DNA (cfDNA) and the evaluation of mutations or methylation pattern. However, expressed RNA can capture mutations, changes in expression levels due to methylation, and provide information on cell of origin, growth, and proliferation status. We developed an approach to isolate cell-free total nucleic acid (cfDNA) and used targeted next generation sequencing to sequence cell-free RNA (cfRNA) and cfDNA as new approach in liquid biopsy. We demonstrate that cfRNA is overall more sensitive than cfDNA in detecting mutations. We show that cfRNA is reliable in detecting fusion genes and cfDNA is reliable in detecting chromosomal gains and losses. cfRNA levels of various solid tumor biomarkers were significantly higher (P < 0.0001) in samples from solid tumors as compared with normal control. Similarly, cfRNA lymphoid markers and cfRNA myeloid markers were all higher in lymphoid and myeloid neoplasms, respectively as compared with control (P < 0.0001). Using machine learning we demonstrate cfRNA was highly predictive of diagnosis (AUC >0.98) of solid tumors, B-cell lymphoid neoplasms, T-cell lymphoid neoplasms, and myeloid neoplasms. In evaluating the host immune system, cfRNA CD4:CD8B and CD3D:CD19 ratios in normal controls were as expected (median: 5.92 and 6.87, respectively) and were significantly lower in solid tumors (P < 0.0002). This data suggests that liquid biopsy combining analysis of cfRNA with cfDNA is practical and may provide helpful information in predicting genomic abnormalities, diagnosis of neoplasms and evaluating both the tumor biology and the host response.
(Methods section) We used Apostle MiniMax High Efficiency cfRNA/cfDNA isolation kit and followed the recommended protocol. After extraction, half of the cfDNA was treated with DNase to obtain cfRNA and the other half was used for DNA studies.
50. Noninvasive Prenatal Screening for Common Fetal Aneuploidies Using Single-Molecule Sequencing. Yeqing Qian, Yongfeng Liu, Kai Yan, et al. Laboratory Investigation Volume 103, Issue 4, April 2023, 100043
(Note: Apostle MiniMax technology is used in this study.)
Amplification biases caused by next-generation sequencing (NGS) for noninvasive prenatal screening (NIPS) may be reduced using single-molecule sequencing (SMS), during which PCR is omitted. Therefore, the performance of SMS-based NIPS was evaluated. We used SMS-based NIPS to screen for common fetal aneuploidies in 477 pregnant women. The sensitivity, specificity, positive predictive value, and negative predictive value were estimated. The GC-induced bias was compared between the SMS- and NGS-based NIPS methods. Notably, a sensitivity of 100% was achieved for fetal trisomy 13 (T13), trisomy 18 (T18), and trisomy 21 (T21). The positive predictive value was 46.15% for T13, 96.77% for T18, and 99.07% for T21. The overall specificity was 100% (334/334). Compared with NGS, SMS (without PCR) had less GC bias, a better distinction between T21 or T18 and euploidies, and better diagnostic performance. Overall, our results suggest that SMS improves the performance of NIPS for common fetal aneuploidies by reducing the GC bias introduced during library preparation and sequencing.
(Materials and Methods section) - cfDNA was extracted from 0.6 mL of maternal plasma using Apostle MiniMaxTM High-Efficiency cfDNA Isolation Kit (ref. number: A17622-50; Apostle) according to the manufacturer’s instructions.
49. Rapid Lineage Assignment of Severe Acute Respiratory Syndrome Coronavirus 2 Cases through Automated Library Preparation, Sequencing, and Bioinformatic Analysis. Andrew J. Gorzalski, Heather Kerwin, Subhash Verma, David C. Hess, Joel Sevinsky, Kevin Libuit, Irina Vlasova-St. Louis, Danielle Siao, Lauren Siao, Diego Buñuel, Stephanie Van Hooser, Mark W. Pandori. The Journal of Molecular Diagnostics Volume 25, Issue 4, April 2023, Pages 191-196
(Note: Apostle MiniGenomics, Apostle MagTouch technologies are used in this study.)
The coronavirus disease 2019 (COVID-19) pandemic has provided a stage to illustrate that there is considerable value in obtaining rapid, whole-genome–based information about pathogens. This article describes the utility of a commercially available, automated severe acute respiratory syndrome associated coronavirus 2 (SARS-CoV-2) library preparation, genome sequencing, and a bioinformatics analysis pipeline to provide rapid, near–real-time SARS-CoV-2 variant description. This study evaluated the turnaround time, accuracy, and other quality-related parameters obtained from commercially available automated sequencing instrumentation, from analysis of continuous clinical samples obtained from January 1, 2021, to October 6, 2021. This analysis included a base-by-base assessment of sequencing accuracy at every position in the SARS-CoV-2 chromosome using two commercially available methods. Mean turnaround time, from the receipt of a specimen for SARS-CoV-2 testing to the availability of the results, with lineage assignment, was <3 days. Accuracy of sequencing by one method was 100%, although certain sites on the genome were found repeatedly to have been sequenced with varying degrees of read error rate.
(Material and Methods section) - Nucleic acid was extracted using the MagTouch Nucleic Acid Extraction Automation System (Apostle Inc., San Jose, CA).
48. Cell-free chromatin immunoprecipitation can determine tumor gene expression in lung cancer patients. Christoffer Trier Maansson, Peter Meldgaard, Magnus Stougaard, Anders Lade Nielsen, Boe Sandahl Sorensen. Molecular Oncology. February 24, 2023. DOI: 10.1002/1878-0261.13394.
(Note: Apostle MiniMax technology is used in this study.)
Cell-free DNA (cfDNA) in blood plasma can be bound to nucleosomes that contain post-translational modifications representing the epigenetic profile of the cell of origin. This includes histone H3 lysine 36 trimethylation (H3K36me3), a marker of active transcription. We hypothesized that cell-free chromatin immunoprecipitation (cfChIP) of H3K36me3-modified nucleosomes present in blood plasma can delineate tumor gene expression levels. H3K36me3 cfChIP followed by targeted NGS (cfChIP-seq) was performed on blood plasma samples from non-small cell lung cancer patients (NSCLC, n = 8), small cell lung cancer patients (SCLC, n = 4) and healthy controls (n = 4). H3K36me3 cfChIP-seq demonstrated increased enrichment of mutated alleles compared to normal alleles in plasma from patients with known somatic cancer mutations. Additionally, genes identified to be differentially expressed in SCLC and NSCLC tumors had concordant H3K36me3 cfChIP enrichment profiles in NSCLC (sensitivity = 0.80) and SCLC blood plasma (sensitivity = 0.86). Findings here expand the utility of cfDNA in liquid biopsies to characterize treatment resistance, cancer subtyping, and disease progression.
(Material and Methods section) -Both the input as well as the cfChIP samples were purified using Apostle MiniMax High Efficiency cfDNA Isolation Kit (Beckman Coulter, Indianapolis, IN, USA) according to manufacturer’s instructions.
47. Automatic Separation and Collection of Cancer-Related Substances from Clinical Samples. Jin-Han Bae, Jay Jeong, Byung Chul Kim, et al. Journal of Visualized Experiments. January 13th, 2023. DOI: 10.3791/64325
(Note: Apostle MiniMax technology is used in this study.)
Recently, liquid biopsies have been used to diagnose various diseases, including cancer. Body fluids contain many substances, including cells, proteins, and nucleic acids originating from normal tissues, but some of these substances also originate from the diseased area. The investigation and analysis of these substances in the body fluids play a pivotal role in the diagnosis of various diseases. Therefore, it is important to accurately separate the required substances, and several techniques are developed to be used for this purpose. We have developed a lab-on-a-disc type of device and platform named CD-PRIME. This device is automated and has good results for sample contamination and sample stability. Moreover, it has advantages of a good acquisition yield, a short operation time, and high reproducibility. In addition, depending on the type of disc to be mounted, plasma containing cell-free DNA, circulating tumor cells, peripheral blood mononuclear cells, or buffy coats can be separated. Thus, the acquisition of a variety of materials present in the body fluids can be done for a variety of downstream applications, including the study of omics.
(Materials section) - Apostle MiniMax High Efficiency Cell-Free DNA Isolation Kit Apostle A17622-250 5 mL X 50 preps version
46. Comparison of cfDNA content in different cancer types with different extraction methods. Wenlong Zhang, Yanan Zhao, Jun Li, et al. Journal of Clinical Oncology 41, no. 16_suppl (June 01, 2023) e15026-e15026.
(Note: Apostle MiniMax technology is used in this study.)
Background: ctDNA liquid biopsy technology has been widely used in the whole course of cancer management, from early screening and early diagnosis of tumors to tumor monitoring and medication guidance. However, the performances of ctDNA as a biomarker in these scenarios are different among different cancer types. One possible reason is that different types of cancer release different amounts of ctDNA or cfDNA, therefore, we explored the cfDNA levels in different cancer types in this study.
Methods: We collected 15913 peripheral blood samples from 14686 cancer patients with different clinicopathologies, 256 patients with pulmonary nodules, and 971 healthy people. We isolated their plasma and extracted the cfDNA using QIAamp Circulating Nucleic Acid Kit, Apostle MiniMax cfDNA Extraction Kit or HaploX magnetic bead method cfDNA extraction kit. We calculated the average concentration of cfDNA (ng/ml) for different groups.
Results: The number of samples of cervical cancer, liver cancer, glioma, colon cancer, prostate cancer, nasopharyngeal carcinoma, gallbladder carcinoma, melanoma, cholangiocarcinoma, colorectal cancer, breast cancer, gastric cancer, endometrial carcinoma, lung cancer, urothelial carcinoma, rectal cancer, ovarian cancer, healthy people, and pulmonary nodule patients are 93, 469, 113, 277, 139, 76, 89, 132, 171, 4010, 407, 547, 68, 7750, 46, 203, 96, 971 and 256, respectively. The cfDNA level of corresponding groups are 88.07, 79.51, 75.57, 63.58, 61.37, 59.09, 57.59, 57.16, 42.67, 34.73, 31.75, 31.37, 31.33, 29.84, 28.72, 28.69, 23.83, 17.82 and 12.54 ng/ml, respectively. The main groups are shown in the table below.
Conclusions: The cfDNA concentrations of healthy people or pulmonary nodule patients were significantly lower than that of tumor patients, and there were also significant differences between different types of cancer. What's interesting, the cfDNA concentrations of lung cancer patients, pulmonary nodule patients and healthy people were quite different. What’s more, there were differences between Colon cancer and Rectal cancer patients. So, the cfDNA concentration maybe a good dimention to be used in early cancer screening and tracing of tumor types.
Publications (2022)
45. Individualized, heterologous chimpanzee adenovirus and self-amplifying mRNA neoantigen vaccine for advanced metastatic solid tumors: phase 1 trial interim results. Christine D. Palmer, Amy R. Rappaport, Matthew J. Davis, et al. Nature Medicine volume 28, pages 1619–1629 (2022)
(Note: Apostle MiniMax technology is used in this study.)
Abstract Checkpoint inhibitor (CPI) therapies provide limited benefit to patients with tumors of low immune reactivity. T cell-inducing vaccines hold promise to exert long-lasting disease control in combination with CPI therapy. Safety, tolerability and recommended phase 2 dose (RP2D) of an individualized, heterologous chimpanzee adenovirus (ChAd68) and self-amplifying mRNA (samRNA)-based neoantigen vaccine in combination with nivolumab and ipilimumab were assessed as primary endpoints in an ongoing phase 1/2 study in patients with advanced metastatic solid tumors (NCT03639714). The individualized vaccine regimen was safe and well tolerated, with no dose-limiting toxicities. Treatment-related adverse events (TRAEs) >10% included pyrexia, fatigue, musculoskeletal and injection site pain and diarrhea. Serious TRAEs included one count each of pyrexia, duodenitis, increased transaminases and hyperthyroidism. The RP2D was 1012 viral particles (VP) ChAd68 and 30 µg samRNA. Secondary endpoints included immunogenicity, feasibility of manufacturing and overall survival (OS). Vaccine manufacturing was feasible, with vaccination inducing long-lasting neoantigen-specific CD8 T cell responses. Several patients with microsatellite-stable colorectal cancer (MSS-CRC) had improved OS. Exploratory biomarker analyses showed decreased circulating tumor DNA (ctDNA) in patients with prolonged OS. Although small study size limits statistical and translational analyses, the increased OS observed in MSS-CRC warrants further exploration in larger randomized studies.
(Methods section) cfDNA was extracted from the entire plasma volume of a single draw using the Apostle MiniMax cfDNA Isolation kit (ApostleBio)
44. Simultaneous analysis of mutations and methylations in circulating cell-free DNA for hepatocellular carcinoma detection. Pei Wang, Qianqian Song, Jie Ren, et al. Science Translational Medicine 14, eabp8704 (2022) 23 November 2022
(Note: Apostle MiniMax technology is used in this study.)
Cell-free DNA (cfDNA)–based liquid biopsy is a promising approach for the early detection of cancer. A major hurdle is the limited yield of cfDNA from one blood draw, limiting the use of most samples to one test of either mutation or methylation. Here, we develop a technology, Mutation Capsule Plus (MCP), which enables multiplex profiling of one cfDNA sample, including simultaneous detection of genetic and epigenetic alterations and genome-wide discovery of methylation markers. With this technology, we performed de novo screening of methylation markers on cfDNA samples from 30 hepatocellular carcinoma (HCC) cases and 30 non-HCC controls. The methylation markers enriched in HCC cfDNA were further profiled in parallel with a panel of mutations on a training cohort of 60 HCC and 60 non-HCC cases, resulting in an HCC detection model. We validated the model in an independent retrospective cohort with 58 HCC and 198 non-HCC cases and got 90% sensitivity with 94% specificity. Furthermore, we applied the model to a prospective cohort of 311 asymptomatic hepatitis B virus carriers with normal liver ultrasonography and serum AFP concentration. The model detected four of the five HCC cases in the cohort, showing 80% sensitivity and 94% specificity. These findings demonstrate that the MCP technology has potential for the discovery and validation of multiomics biomarkers for the noninvasive detection of cancer. This study also provides a comprehensive database of genetic and epigenetic alterations in the cfDNA of a large cohort of HCC cases and high-risk non-HCC individuals.
(Methods Section) cfDNA was extracted from the plasma samples using the Apostle MiniMax cfDNA isolation kit (C43468, Apostle).
43. Safety and tolerability of AAV8 delivery of a broadly neutralizing antibody in adults living with HIV: a phase 1, dose-escalation trial. Casazza JP, Cale EM, et al. the VRC603 Study Team. Nature Medicine April 11, 2022; https://www.nature.com/articles/s41591-022-01762-x
(Note: Apostle MiniMax technology is used in this study.)
Adeno-associated viral vector-mediated transfer of DNA coding for broadly neutralizing anti-HIV antibodies (bnAbs) offers an alternative to attempting to induce protection by vaccination or by repeated infusions of bnAbs. In this study, we administered a recombinant bicistronic adeno-associated virus (AAV8) vector coding for both the light and heavy chains of the potent broadly neutralizing HIV-1 antibody VRC07 (AAV8-VRC07) to eight adults living with HIV. All participants remained on effective anti-retroviral therapy (viral load (VL) <50 copies per milliliter) throughout this phase 1, dose-escalation clinical trial (NCT03374202). AAV8-VRC07 was given at doses of 5 × 1010, 5 × 1011 and 2.5 × 1012 vector genomes per kilogram by intramuscular (IM) injection. Primary endpoints of this study were to assess the safety and tolerability of AAV8-VRC07; to determine the pharmacokinetics and immunogenicity of in vivo VRC07 production; and to describe the immune response directed against AAV8-VRC07 vector and its products. Secondary endpoints were to assess the clinical effects of AAV8-VRC07 on CD4 T cell count and VL and to assess the persistence of VRC07 produced in participants. In this cohort, IM injection of AAV8-VRC07 was safe and well tolerated. No clinically significant change in CD4 T cell count or VL occurred during the 1–3 years of follow-up reported here. In participants who received AAV8-VRC07, concentrations of VRC07 were increased 6 weeks (P = 0.008) and 52 weeks (P = 0.016) after IM injection of the product. All eight individuals produced measurable amounts of serum VRC07, with maximal VRC07 concentrations >1 µg ml−1 in three individuals. In four individuals, VRC07 serum concentrations remained stable near maximal concentration for up to 3 years of follow-up. In exploratory analyses, neutralizing activity of in vivo produced VRC07 was similar to that of in vitro produced VRC07. Three of eight participants showed a non-idiotypic anti-drug antibody (ADA) response directed against the Fab portion of VRC07. This ADA response appeared to decrease the production of serum VRC07 in two of these three participants. These data represent a proof of concept that adeno-associated viral vectors can durably produce biologically active, difficult-to-induce bnAbs in vivo, which could add valuable new tools to the fight against infectious diseases.
(Methods Section) AAV8-VRC07 vector DNA quantitation. Plasma AAV8-VRC07 plasmid DNA was measured by extracting DNA from plasma, concentrating and then using a real-time PCR assay to measure a 103 base sequence spanning the junction of the IgG heavy chain sequence and F2A insert. DNA was extracted from serum using an Apostle MiniMax High Efficiency cfDNA Isolation Kit, following the manufacturer’s protocol with slight modification.
42. Comparative analysis of cell-free DNA extraction efficiency from plasma. Beckman Coulter Life Sciences. Indianapolis, IN. Technical Note 2022.
(Note: Apostle MiniMax technology is compared and discussed in this study.)

Results and discussion
Mean cfDNA concentrations in the eluates were determined to be 8.30 ng/μL for samples extracted with the QIAamp kit and 11.15 ng/μL for samples extracted with the Apostle kit when measured with the Qubit assay (Figure 1A). This illustrates a 34.3% higher yield when using the Apostle kit as compared to the QIAamp kit.
When evaluating the contents of specific genes in the eluate, the Apostle purification displayed a higher yield than QIAamp (Figure 1B). 8358 DNAJC5G positive droplets were detected in the Apostle samples compared to 7269 droplets in the QIAamp samples. Similarly, 10849 ACTG1 droplets were detected in the Apostle-extracted samples versus 9463 droplets detected in the QIAamp-extracted samples. This data demonstrates an average of 14.8% higher yield using the Apostle kit.
Finally, when evaluating cfDNA fragments via the Agilent bioanalyzer instrument, the Apostle kit demonstrated the highest yield (Figure 1C and D). Mononucleosomal cfDNA concentration in the eluate of Apostle samples was estimated to be 10.64 ng/μL versus 6.26 ng/μL in the eluate of QIAamp samples. The dinucleosomal cfDNA concentration was low for both kits, with 0.13 ng/μL recorded for Apostle samples and 0.24 ng/μL for QIAamp samples. This indicates a slightly better purification of dinucleosomal cfDNA using QIAamp; however, this could also be an artifact given the low concentrations. The Apostle mononucleosomal cfDNA yield was 70.0% higher than the yield for the QIAamp samples.

A summary provided in Table 2 shows the consistently higher yield achieved by the Apostle MiniMax High Efficiency cfDNA Isolation Kit as compared to the QIAamp® circulating nucleic acid kit. The average 40% improvement in yield underscores the importance of extraction kit selection in cfDNA assay development and highlights the potential impact of cfDNA isolation efficiency in ctDNA detection.
41. Reliability of Cell-Free DNA and Targeted NGS in Predicting Chromosomal Abnormalities of Patients With Myeloid Neoplasms. Andrew Ip, Alexandra Della Pia, Gee Youn (Geeny) Kim, Jason Lofters, James Behrmann, Dylon Patel, Simone Kats, Jeffrey Justin Estella, Ivan De Dios, Wanlong Ma, Andrew L. Pecora, Andre H. Goy, Jamie Koprivnikar, James K. McCloskey and Maher Albitar. Frontiers in Oncology June 14, 2022; https://doi.org/10.3389/fonc.2022.923809
(Note: Apostle MiniMax technology is used in this study.)
Introduction: Cytogenetic analysis is important for stratifying patients with various neoplasms. We explored the use of targeted next generation sequencing (NGS) in detecting chromosomal structural abnormalities or copy number variations (CNVs) in patients with myeloid neoplasms.
Methods: Plasma cell-free DNA (cfDNA) from 2821 myeloid or lymphoid neoplasm patients were collected. cfDNA was sequenced using a 275 gene panel. CNVkit software was used for analyzing and visualizing CNVs. Cytogenetic data from corresponding bone marrow (BM) samples was available on 89 myeloid samples.
Results: Of the 2821 samples, 1539 (54.5%) showed evidence of mutations consistent with the presence of neoplastic clones in circulation. Of these 1539 samples, 906 (59%) showed abnormalities associated with myeloid neoplasms and 633 (41%) with lymphoid neoplasms. Chromosomal structural abnormalities in cfDNA were detected in 146 (16%) myeloid samples and 76 (12%) lymphoid samples. Upon comparison of the myeloid samples with 89 BM patients, NGS testing was able to reliably detect chromosomal gain or loss, except for fusion abnormalities. When cytogenetic abnormalities were classified according to prognostic classes, there was a complete (100%) concordance between cfDNA NGS data and cytogenetic data.
Conclusions: This data shows that liquid biopsy using targeted NGS is reliable in detecting chromosomal structural abnormalities in myeloid neoplasms. In specific circumstances, targeted NGS may be reliable and efficient to provide adequate information without the need for BM biopsy considering broad mutation profiling can be obtained through adequate sequencing within the same test. Overall, this study supports the use of liquid biopsy for early diagnosis and monitoring of patients with myeloid neoplasms.
(Methods Section) Sample Collection and cfDNA Isolation
We extracted cfDNA from 2821 peripheral blood samples using the Apostle MiniMax(TM) High Efficiency cfDNA Isolation Kit (San Jose, CA). These peripheral blood samples were collected in EDTA anticoagulant over approximately 18 months. DNA was extracted from separated plasma within 48 hours of collection. We tested these samples for the presence of circulating tumor DNA (ctDNA).
40. Quantifying Fetal DNA in Maternal Blood Plasma by ddPCR Using DNA Methylation. E. Hall, T. Riel, M. Ramesh, M. Gencoglu, O. Mikhaylichenko, R. Dannebaum, S. Margeridon, M. Herrera. Bio-Rad Laboratories, Pleasanton, CA. Association for Molecular Pathology 2022 Annual Meeting Abstracts. J Mol Diagn 2022, 24:S1 Abstract G072. (Full Abstract List) (G072 Only)
(Note: Apostle MiniMax technology is used in this study.)
Introduction: The proportion of cell-free DNA (cfDNA) circulating in maternal blood that originates from the fetus, the fetal fraction, is an important quality control metric when performing tests on fetal-derived cfDNA. Epigenetic differences produce dissimilar DNA methylation patterns, allowing for leveraging regions of high methylation contrast using methylation-sensitive restriction enzyme (MSRE) digestion to quantify fetal and maternal DNA via droplet digital PCR (ddPCR). This advancement positions ddPCR as a faster and less expensive alternative to next-generation sequencing (NGS) for fetal fraction estimation.
Methods: Assays were designed to target MSRE- compatible regions with high methylation contrast between maternal and fetal cfDNA. Fetal assays targeted sites hypermethylated in fetal cfDNA and maternal assays targeted sites hypermethylated in maternal cfDNA. The assay multiplex was tested against contrived and clinical samples using an in-droplet MSRE-ddPCR workflow. The reaction mix was dropletized to create about 20,000 droplets per 24-μL reaction, thermocycled, and analyzed in a QX ONE instrument. The thermocycling profile included a 45-minute MSRE incubation step prior to PCR amplification. Contrived samples were constructed by spiking DNA-free plasma with micrococcal nuclease-digested DNA from an amniotic fluid cell line ("fetal" component) and a B-lymphocyte cell line ("maternal" component). Clinical samples were remnant diagnostic samples with existing NGS non-invasive prenatal testing results attached. DNA was extracted from all samples with the Apostle MiniMax kit on the KingFisher Flex.
Results: From an initial set of 15 assays, a final five-assay multiplex was produced following amplicon sequencing with NGS and ddPCR screening. Although amplicon sequencing did not completely predict ddPCR performance and non- specific interactions, it was valuable for guiding the final ddPCR screen. The five-assay multiplex, consisting of three fetal assays and two maternal assays, produced an excellent linear response against contrived samples from 0% to 25% fetal fraction (R2 >0.99). Similarly, a high correlation was observed between ddPCR-estimated fetal fraction and NGS fetal fraction for a set of clinical samples (n=6, plus two non- pregnant controls, R2 >0.94).
Conclusions: As an epigenetic trait, DNA methylation is a useful way to discriminate between otherwise highly similar DNA sequences in an efficient and effective manner. Leveraging DNA methylation may be done with minimal impact to the standard ddPCR workflow. The high sensitivity, speed, and direct quantification of ddPCR make it an attractive alternative to NGS for fetal fraction estimation.
39. Next-Generation Digital Polymerase Chain Reaction: High-Dynamic-Range Single-Molecule DNA Counting via Ultrapartitioning. Eleen Y. Shum, Janice H. Lai, Sixing Li, Haeun G. Lee, Jesse Soliman, Vedant K. Raol, Cavina K. Lee, Stephen P.A. Fodor, H. Christina Fan Anal. Chem. 2022, Publication Date:December 12, 2022 https://doi.org/10.1021/acs.analchem.2c03649
(Note: Apostle MiniMax technology is used in this study.)
ABSTRACT: Digital PCR (dPCR) was first conceived for single-molecule quantitation. However, current dPCR systems often require DNA templates to share partitions due to limited partitioning capacities. Here, we introduce UltraPCR, a next-generation dPCR system where DNA counting is performed in a single-molecule regimen through a 6-log dynamic range using a swift and parallelized workflow. Each UltraPCR reaction is divided into >30 million partitions without microfluidics to achieve single template occupancy. Combined with a unique emulsion chemistry, partitions are optically clear, enabling the use of a three-dimensional imaging technique to rapidly detect DNA-positive partitions. Single-molecule occupancy also allows for more straightforward multiplex assay development due to the absence of partition-specific competition. As a proof of concept, we developed a 222-plex UltraPCR assay and demonstrated its potential use as a rapid, low-cost screening assay for noninvasive prenatal testing for as low as 4% trisomy fraction samples with high precision, accuracy, and reproducibility.
(Methods Section) Cell-free plasma was isolated using the Apostle MiniMax kit (Beckman Coulter) according to the manufacturer’s protocol with an elution volume of 60 μL.
38. A method for early diagnosis of lung cancer from tumor originated DNA fragments using plasma cfDNA methylome and fragmentome profiles. Yeo Jin Kim, Hahyeon Jeona, Sungwon Jeon, et al. Molecular and Cellular Probes Volume 66, December 2022
(Note: Apostle MiniMax technology is used in this study.)
Abstract
Early detection is critical for minimizing mortality from cancer. Plasma cell-free DNA (cfDNA) contains the signatures of tumor DNA, allowing us to quantify the signature and diagnose early-stage tumors. Here, we report a novel tumor fragment quantification method, TOF (Tumor Originated Fragment) for the diagnosis of lung cancer by quantifying and analyzing both the plasma cfDNA methylation patterns and fragmentomic signatures. TOF utilizes the amount of ctDNA predicted from the methylation density information of each cfDNA read mapped on 6243 lung-tumor-specific CpG markers. The 6243 tumor-specific markers were derived from lung tumor tissues by comparing them with corresponding normal tissues and healthy blood from public methylation data. TOF also utilizes two cfDNA fragmentomic signatures: 1) the short fragment ratio, and 2) the 5′ end-motif profile. We used 298 plasma samples to analyze cfDNA signatures using enzymatic methyl-sequencing data from 201 lung cancer patients and 97 healthy controls. The TOF score showed 0.98 of the area under the curve in correctly classifying lung cancer from normal samples. The TOF score resolution was high enough to clearly differentiate even the early-stage non-small cell lung cancer patients from the healthy controls. The same was true for small cell lung cancer patients.
(Methods Section) Cell-free DNA was extracted from 3 to 5 ml of plasma using XXX method or Apostle MiniMaxTM High Efficiency Isolation Kit (Beckman Coulter Life Sciences, C40603), according to the manufacturers’ procedures.
37. Nicotine dose-dependent epigenomic-wide DNA methylation changes in the mice with long-term electronic cigarette exposure. Gang Peng,* Yibo Xi,* Chiara Bellini, Kien Pham, Zhen W Zhuang, Qin Yan, Man Jia, Guilin Wang, Lingeng Lu, Moon-Shong Tang, Hongyu Zhao, and He Wang American Journal of Cancer Research. Volume 12, Issue 8, August 2022, Pages 3679–3692.
(Note: Apostle MiniMax technology is us in this study.)
Abstract
Epigenomic-wide DNA methylation profiling holds the potential to reflect both electronic cigarette exposure-associated risks and individual poor health outcomes. However, a systemic study in animals or humans is still lacking. Using the Infinium Mouse Methylation BeadChip, we examined the DNA methylation status of white blood cells in male ApoE-/- mice after 14 weeks of electronic cigarette exposure with the InExpose system (2 hr/day, 5 days/week, 50% PG and 50% VG) with low (6 mg/ml) and high (36 mg/ml) nicotine concentrations. Our results indicate that electronic cigarette aerosol inhalation induces significant alteration of 8,985 CpGs in a dose-dependent manner (FDR<0.05); 7,389 (82.2%) of the CpG sites are annotated with known genes. Among the top 6 significant CpG sites (P-value<1e-8), 4 CpG sites are located in the known genes, and most (3/5) of these genes have been related to cigarette smoking. The other two CpGs are close to/associated with the Phc2 gene that was recently linked to smoking in a transcriptome-wide associations study. Furthermore, the gene set enrichment analysis highlights the activation of MAPK and 4 cardiomyocyte/cardiomyopathy-related signaling pathways (including adrenergic signaling in cardiomyocytes and arrhythmogenic right ventricular cardiomyopathy) following repeated electronic cigarette use. The MAPK pathway activation correlates well with our finding of increased cytokine mRNA expression after electronic cigarette exposure in the same batch of mice. Interestingly, two pathways related to mitochondrial activities, namely mitochondrial gene expression and mitochondrial translation, are also activated after electronic cigarette exposure. Elucidating the relationship between these pathways and the increased circulating mitochondrial DNA observed here will provide further insight into the cell-damaging effects of prolonged inhalation of e-cigarette aerosols.
(Methods section) Plasma cfDNA and mtDNA/nDNA ratio and oxidization of cell-free DNA
For measurement of in vivo plasma mtDNA/nDNA ratio, mice plasma was first subjected to cell-free DNA (cfDNA) extraction using Apostle minimax cfDNA extraction kit (ApostleBio, CA, US).
36. High-throughput sample processing for methylation analysis in an automated, enclosed environment. Alejandro Stark, Thomas R. Pisanic, James G. Herman, Tza-Huei Wang. SLAS Technology Volume 27, Issue 3, June 2022, Pages 172-179.
(Note: Apostle MiniMax technology is compared and discussed in this study.)

(Figure 4) - "Most notably, the Apostle particles outperformed all others, achieving almost 2-fold higher recovery yields than the particles supplied in the X kit. "
Variation in methylcytosine is perhaps the most well-studied epigenetic mechanism of gene regulation. Methods that have been developed and implemented for assessing DNA methylation require sample DNA to be extracted, purified and chemically-processed through bisulfite conversion before downstream analysis. While some automated solutions exist for each of these individual process steps, a fully integrated solution for accomplishing the entire process in a high-throughput manner has yet to be demonstrated. Thus, sample processing methods still require numerous manual steps that may reduce sample throughput and precision, while increasing the risk of contamination and human error. In this work, we present an integrated, automated solution for performing the entire sample preparation process, including DNA extraction, purification, bisulfite conversion and PCR plate preparation within in an enclosed environment. The method employs silica-coated magnetic particles that eliminate the need for a centrifuge or vacuum manifold, thereby reducing the complexity and cost of the required automation platform. Toward this end, we also compare commercial DNA extraction and bisulfite conversion kits to identify a protocol suitable for automation to significantly improve genomic and bisulfite-treated DNA yields over manufacturer protocols. Overall, this research demonstrated development of an automated protocol that offers the ability to generate high-quality, bisulfite-treated DNA samples in a high-throughput and clean environment with minimal user intervention and comparable yields to manual processing.
35. Detecting SARS-CoV-2 variants in wastewater and their correlation with circulating variants in the communities. Lin Li, Timsy Uppal, Paul D. Hartley, Andrew Gorzalski, Mark Pandori, Michael A. Picker, Subhash C. Verma & Krishna Pagilla. Scientific Reports volume 12, Article number: 16141 (2022) , September 2022
(Note: Apostle MiniGenomics technology is used in this study.)
Abstract
Detection of SARS-CoV-2 viral load in wastewater has been highly informative in estimating the approximate number of infected individuals in the surrounding communities. Recent developments in wastewater monitoring to determine community prevalence of COVID-19 further extends into identifying SARS-CoV-2 variants, including those being monitored for having enhanced transmissibility. We sequenced genomic RNA derived from wastewater to determine the variants of coronaviruses circulating in the communities. Wastewater samples were collected from Truckee Meadows Water Reclamation Facility (TMWRF) from November 2020 to June 2021. SARS-CoV-2 variants resulting from wastewater were compared with the variants detected in infected individuals' clinical specimens (nasal/nasopharyngeal swabs) during the same period and found conclusively in agreement. Therefore, wastewater monitoring for SARS-CoV-2 variants in the community is a feasible strategy as a complementary tool to clinical specimen testing in the latter's absence.
(Methods Section) The NP swabs received at the Nevada State Public Health Laboratory (NSPHL) from the Reno-Sparks metropolitan for SARS-CoV-2 diagnostic testing were subjected for RNA extraction using Virus Total NA Isolation Fast Kit (Apostle MiniGenomics, San Jose, CA, USA), followed by RT-PCR for the detection of SARS-CoV-2 genome using US Federal Drug Administration Emergency Use Authorization (FDA-EUA) diagnostic kits, as described previously.
34. Optimalizace digitální polymerázové řetězové reakce pro aplikaci v neinvazivní prenatální diagnostice (Optimization of digital polymerase chain reaction for application in non-invasive prenatal diagnostics) Author: Šenkyřík, Pavel; Advisor: Korabečná, Marie; Referee: Vodička, Radek; Faculty / Institute: Faculty of Science; Discipline: Anthropology and Human Genetics; Department: Department of Anthropology and Human Genetics; Date of defense: 13. 9. 2022; Publisher: Univerzita Karlova, Přírodovědecká fakulta; Language: Czech
(Note: Apostle MiniMax technology is discussed in this diploma thesis.)
(Note: Table 13 referenced below in this diploma thesis shows that Apostle MiniMax yields the greatest number of analyzable drops in the digital PCR.)

33. Rapid repeat infection of SARS-CoV-2 by two highly distinct delta-lineage viruses. Andrew J. Gorzalski , Christina Boyles, Victoria Sepcic, Subhash Verma, Joel Sevinsky, Kevin Libuit, Stephanie Van Hooser, Mark W. Pandori. Diagnostic Microbiology and Infectious Disease Volume 104, Issue 1, September 2022, 115747; https://doi.org/10.1016/j.diagmicrobio.2022.115747
(Note: Apostle MagTouch technology is used in this study.)
An instance of sequential infection of an individual with, firstly, the Delta variant and secondly a Delta-sub-lineage has been identified. The individual was found positive for the AY.26 lineage 22 days after being found positive for the Delta [B.1.617.2] variant. The viruses associated with the cases showed dramatic genomic difference, including 31 changes that resulted in deletions or amino acid substitutions. Seven of these differences were observed in the Spike protein. The patient in question was between 30 and 35 years old and had no underlying health conditions. Though singular, this case illustrates the possibility that infection with the Delta variant may not itself be fully protective against a population of SARS-CoV-2 variants that are becoming increasingly diverse.
32. Response prediction and risk stratification of patients with rectal cancer after neoadjuvant therapy through an analysis of circulating tumour DNA. Liu W, Li Y, et al. EBioMedicine March 17, 2022; https://www.thelancet.com/journals/ebiom/article/PIIS2352-3964(22)00129-3/fulltext
(Note: Apostle MiniMax technology is used in this study.)
Background - Multiple approaches based on cell-free DNA (cfDNA) have been applied to detect minimal residual disease (MRD) and to predict prognosis or recurrence. However, a comparison of the approaches used in different cohorts and studies is difficult. We aimed to compare multiple approaches for MRD analysis after neoadjuvant therapy (NAT) in patients with locally advanced rectal cancer (LARC).
Methods - Sixty patients with LARC from a multicentre, phase II/III randomized trial were included, with tissue and blood samples collected. For each cfDNA sample, we profiled MRD using 3 approaches: personalized assay targeting tumour-informed mutations, universal panel of genes frequently mutated in colorectal cancer (CRC), and low depth sequencing for copy number alterations (CNAs).
Findings - Positive MRD based on post-NAT personalized assay was significantly associated with an increased risk of recurrence (HR = 27.38; log-rank P < 0.0001). MRD analysis based on universal panel (HR = 5.18; log-rank P = 0.00086) and CNAs analysis (HR = 9.24; log-rank P = 0.00017) showed a compromised performance in predicting recurrence. Both the personalized assay and universal panel showed complementary pattern to CNAs analysis in detecting cases with recurrence and the combination of the two types of biomarkers may lead to better performance.
Interpretation - The combination of mutation profiling and CNA profiling can improve the detection of MRD, which may help optimize the treatment strategies for patients with LARC.
31. A microfluidic platform for high-purity cell free DNA extraction from plasma for non-invasive prenatal testing. Lindsay Schneider, Thomas Usherwood, Anubhav Tripathi. Prenatal Diagnosis January 14, 2022; https://doi.org/10.1002/pd.6092
(Note: Apostle MiniMax technology is discussed in this study.)
Objectives
Increase the yield and purity of cell-free DNA (cfDNA) extracted from plasma for non-invasive prenatal testing (NIPT) as inefficiencies in this extraction and purification can dramatically affect the sensitivity and specificity of the test.
Methods
This work integrates cfDNA extraction from plasma with a microfluidic chip platform by combining magnetic bead-based extraction and electroosmotic flow on the microfluidic chip. Various wash buffers and voltage conditions were simulated using COMSOL Multiphysics Modeling and tested experimentally.
Results
When performing the first wash step of this assay on the microfluidic chip with 300 V applied across the channel there was a six-fold increase in the A260/A230 ratio showing a significant improvement (p value 0.0005) in the purity of the extracted sample all while maintaining a yield of 68.19%. These values are critical as a high yield results in more sample to analyze and an increase in A260/A230 ratio corresponds to a decrease in salt contaminants such as guanidinium thiocyanate which can interfere with downstream processes during DNA library preparation and potentially hinder the NIPT screening results.
Conclusions
This technique has the potential to improve NIPT outcomes and other clinically relevant workflows that use cfDNA as an analyte such as cancer detection.
30. Direct capture and sequencing reveal ultra-short single-stranded DNA in biofluids. Cheng et al. iScience. 2022; In Press.
(Note: Apostle MiniMax technology is discussed in this study.)
Summary
Cell-free DNA (cfDNA) has become the predominant analyte of liquid biopsy; however, recent studies suggest the presence of subnucleosomal-sized DNA fragments in circulation that are likely single-stranded. Here, we report a method called Direct Capture and Sequencing (DCS) tailored to recover such fragments from biofluids by directly capturing them using short degenerate probes followed by single strand-based library preparation and next generation sequencing. DCS revealed a new DNA population in biofluids, named ultra-short single-stranded DNA (ussDNA). Evaluation of the size distribution and abundance of ussDNA manifested generality of its presence in human, animal species, and plants. In human, red blood cells were found to contain abundant ussDNA; plasma-derived ussDNA exhibited modal size at 50nt. This work reports the presence of an understudied DNA population in circulation, and yet more work is awaiting to study its generation mechanism, tissue of origin, disease implications, etc.
29. Nanostructures in non-invasive prenatal genetic screening. Samira Sadeghi, Mahdi Rahaie & Bita Ostad-Hasanzadeh. Biomedical Engineering Letters volume 12, pages 3–18 (2022)
(Note: Apostle MiniEnrich technology is reviewed in this review article.)
Abstract
Prenatal screening is an important issue during pregnancy to ensure fetal and maternal health, as well as preventing the birth of a defective fetus and further problems such as extra costs for the family and society. The methods for the screening have progressed to non-invasive approaches over the recent years. Limitations of common standard screening tests, including invasive sampling, high risk of abortion and a big delay in result preparation have led to the introduction of new rapid and non-invasive approaches for screening. Non-invasive prenatal screening includes a wide range of procedures, including fetal cell-free DNA analysis, proteome, RNAs and other fetal biomarkers in maternal serum. These biomarkers require less invasive sampling than usual methods such as chorionic villus sampling, amniocentesis or cordocentesis. Advanced strategies including the development of nanobiosensors and the use of special nanoparticles have provided optimization and development of NIPS tests, which leads to more accurate, specific and sensitive screening tests, rapid and more reliable results and low cost, as well. This review discusses the specifications and limitations of current non-invasive prenatal screening tests and introduces a novel collection of detection methods reported studies on nanoparticles’ aided detection. It can open a new prospect for further studies and effective investigations in prenatal screening field.
(pp. 13) In another study, Zhang, et al. designed a magnetic nano-platform named MiniEnrich, used to enrich the cff-DNA fragments in maternal plasma with high resolution by developing two different magnetic nanoparticle types. It was suggested that the mentioned platform had a fair potential to help detect monogenic disorders in non-invasive prenatal screening [108].
28. Monitoring and Identifying Insulin-Like Growth Factor 1 Variants by Liquid Chromatography–High-Resolution Mass Spectrometry in a Clinical Laboratory. Ievgen Motorykin, Allison Li & Zengru Wu. Clinical Applications of Mass Spectrometry in Biomolecular Analysis pp 239–251. (Book chapter) 2022. Part of the Methods in Molecular Biology book series (MIMB,volume 2546)
(Note: Apostle MiniMax technology is used in this study.)
Publications (2021)
27. Segmental duplication as potential biomarkers for non-invasive prenatal testing of aneuploidies. Xinwen Chen, Yifan Li, Qiuying Huang, Xingming Lin, Xudong Wang, Yafang Wang, Ying Liu, Qiushun He, Yinghua Liu, Ting Wang, Zhi-Liang Ji, Qingge Li. EBioMedicine August 11, 2021; https://www.thelancet.com/journals/ebiom/article/PIIS2352-3964(21)00328-5/fulltext
(Note: Apostle MiniMax technology is used in this study.)
We developed a computational program whereby available SD regions can be processed and analyzed efficiently for their potential use as biomarkers of the aneuploidy of interest. For the five common aneuploidies, i.e., trisomy 13, 18, 21, and two sex chromosome aneuploidies, a total of 21,772 candidate SD biomarker sequences together with their corresponding primer/probe sets were generated. The primer/probe sets were tested using a real-time PCR-based multicolour melting curve analysis for simultaneous detection of the five common aneuploidies, and yielded 100% clinical sensitivity and 99.64% specificity when subjected to a clinical evaluation. Following the observations that the SD biomarkers for aneuploidy could be better detected by digital PCR with improved accuracy, we established a noninvasive prenatal testing protocol for trisomy 21 and attained 100% concordance with next generation sequencing.
Our study confirmed that SD regions are preferred biomarkers for aneuploidy detection and in particular SD-based digital PCR could find potential use for NIPT of trisomy. A similar strategy can be applied to other chromosomal abnormality and genetic disorders.
26. Isotopic Peak Index, Relative Retention Time, and Tandem MS for Automated High Throughput IGF-1 Variants Identification in a Clinical Laboratory. Ievgen Motorykin, Hua Li, Nigel J. Clarke, Michael J. McPhaul, and Zengru Wu. Analytical Chemistry August 2021. https://doi.org/10.1021/acs.analchem.1c02566
(Note: Apostle MiniMax technology is used in this study.)
Measuring insulin-like growth factor-1 (IGF-1) is useful for assessing and managing growth-related disorders, such as acromegaly and growth hormone deficiency. High-resolution liquid chromatography–mass spectrometry (LC–MS) is used for measuring IGF-1 due to its molecular specificity, quantitative performance, well-characterized reference materials, and detailed age/sex-specific reference intervals. However, polymorphisms in the IGF1 gene may cause mass shifts in the polypeptide, which can impede quantitation and cause errors in clinical interpretation. We (1) developed a concept of “isotopic peak index”, which allows simultaneous monitoring of 15 IGF-1 variants by using only four m/z ratios; (2) developed a “relative retention time” parameter that allows distinction of previously unresolved variants; and (3) utilized tandem mass spectrometry (MS/MS) to distinguish between the most common pair of variants: isobaric A67T and A70T. All methods were validated with DNA sequencing. This approach identified six variants from the ExAC database, P66A, A67S, S34N, A38 V, A67T, and A70T; two previously reported V44M and A67V variants; and discovered six unreported variants, Y31H, S33P, R50Q, R56K, T41I, and A62T. Major improvements in our workflow include enhanced automation, avoiding detailed manual calculations that are prone to human error, and the ability to monitor more, and discover new, IGF-1 variants. The workflow provides a profile of a patient’s IGF-1 status and can be used to explore genotype–phenotype relationships in IGF-1 variants.
25. FDA EUA Summary: Fulgent COVID-19 by RT-PCR TEST(FULGENT THERAPEUTICS). For In vitro Diagnostic Use. Rx Only. For use under Emergency Use Authorization (EUA) only. US FDA. April 12, 2021.
(Note: Apostle RNA Extraction Method is included in this US FDA EUA.)
Weblink for the US FDA EUA letter: https://www.fda.gov/media/138147/download
24. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy. Shengnan Jin, Dewen Zhu, Fanggui Shao, Shiliang Chen, Ying Guo, Kuan Li, Yourong Wang, Rongxiu Ding, Lingjia Gao, Wen Ma, Tong Lu, Dandan Li, Zhengzheng Zhang, Suili Cai, Xue Liang, Huayu Song, Ling Ji, Jinlei Li, Zhihai Zheng, Feizhao Jiang, Xiaoli Wu, Ju Luan, Huxiang Zhang, Zhengquan Yang, Charles R. Cantor, Chang Xu, and Chunming Ding. PNAS February 2, 2021 118 (5) e2017421118; https://doi.org/10.1073/pnas.2017421118
(Note: Apostle MiniMax technology is used in this study.)
Multiplex assays, involving the simultaneous use of multiple circulating tumor DNA (ctDNA) markers, can improve the performance of liquid biopsies so that they are highly predictive of cancer recurrence. We have developed a single-tube methylation-specific quantitative PCR assay (mqMSP) that uses 10 different methylation markers and is capable of quantitative analysis of plasma samples with as little as 0.05% tumor DNA. In a cohort of 179 plasma samples from colorectal cancer (CRC) patients, adenoma patients, and healthy controls, the sensitivity and specificity of the mqMSP assay were 84.9% and 83.3%, respectively. In a head-to-head comparative study, the mqMSP assay also performed better for detecting early-stage (stage I and II) and premalignant polyps than a published SEPT9 assay. In an independent longitudinal cohort of 182 plasma samples (preoperative, postoperative, and follow-up) from 82 CRC patients, the mqMSP assay detected ctDNA in 73 (89.0%) of the preoperative plasma samples. Postoperative detection of ctDNA (within 2 wk of surgery) identified 11 of the 20 recurrence patients and was associated with poorer recurrence-free survival (hazard ratio, 4.20; P = 0.0005). With subsequent longitudinal monitoring, 14 patients (70%) had detectable ctDNA before recurrence, with a median lead time of 8.0 mo earlier than seen with radiologic imaging. The mqMSP assay is cost-effective and easily implementable for routine clinical monitoring of CRC recurrence, which can lead to better patient management after surgery.
23. A systematic evaluation of stool DNA preparation protocols for colorectal cancer screening via analysis of DNA methylation biomarkers. Shengnan Jin, Qian Ye, Yanping Hong, Wenqing Dai, Chengliang Zhang, Weihao Liu, Ying Guo, Dewen Zhu, Zhengzheng Zhang, Shiliang Chen, Yourong Wang, Dandan Li, Wen Ma, Zhengquan Yang, Jinlei Li, Zhihai Zheng, Ju Luan, Xiaoli Wu, Feizhao Jiang, Chang Xu and Chunming Ding. Clinical Chemistry and Laboratory Medicine (CCLM). 2021; 59(1): 91–99
Objectives
Colorectal cancer (CRC) screening using stool samples is now in routine use where tumor DNA methylation analysis for leading markers such as NDRG4 and SDC2 is an integral part of the test. However, processing stool samples for reproducible and efficient extraction of human genomic DNA remains a bottleneck for further research into better biomarkers and assays.
Methods
We systematically evaluated several factors involved in the processing of stool samples and extraction of DNA. These factors include: stool processing (solid and homogenized samples), preparation of DNA from supernatant and pellets, and DNA extraction with column and magnetic beads-based methods. Furthermore, SDC2 and NDRG4 methylation levels were used to evaluate the clinical performance of the optimal protocol.
Results
The yield of total and human genomic DNA (hgDNA) was not reproducible when solid stool scraping is used, possibly due to sampling variations. More reproducible results were obtained from homogenized stool samples. Magnetic beads-based DNA extraction using the supernatant from the homogenized stool was chosen for further analysis due to better reproducibility, higher hgDNA yield, lower non-hgDNA background, and the potential for automation. With this protocol, a combination of SDC2 and NDRG4 methylation signals with a linear regression model achieved a sensitivity and specificity of 81.82 and 93.75%, respectively.
Conclusions
Through the systematic evaluation of different stool processing and DNA extraction methods, we established a reproducible protocol for analyzing tumor DNA methylation markers in stool samples for colorectal cancer screening.
(Methods section) For magnetic beads-based method, Apostle Stool gDNA Isolation Kit (APOSTLE) was used according to the manufacturer’s instructions. Either 0.2 g pellets or 0.2 mL supernatant from homogenized stool was mixed with 1 mL lysis buffer (APOSTLE) for DNA extractions.
22. Electronic Cigarettes Induce Mitochondrial DNA Damage and Trigger TLR9 (Toll-Like Receptor 9)-Mediated Atherosclerosis. Jieliang Li , Luong Huynh , William D. Cornwell , Moon-Shong Tang , Hannah Simborio , Jing Huang , Beata Kosmider , Thomas J. Rogers , Huaqing Zhao , Michael B. Steinberg , Le Thu Thi Le , Lanjing Zhang , Kien Pham , Chen Liu , He Wang. Arteriosclerosis, Thrombosis, and Vascular Biology. 2021;41:839–853
Objective:
Electronic cigarette (e-cig) use has recently been implicated in promoting atherosclerosis. In this study, we aimed to investigate the mechanism of e-cig exposure accelerated atherosclerotic lesion development.
Approach and Results:
Eight-week-old ApoE−/− mice fed normal laboratory diet were exposed to e-cig vapor (ECV) for 2 hours/day, 5 days/week for 16 weeks. We found that ECV exposure significantly induced atherosclerotic lesions as examined by Oil Red O staining and greatly upregulated TLR9 (toll-like receptor 9) expression in classical monocytes and in the atherosclerotic plaques, which the latter was corroborated by enhanced TLR9 expression in human femoral artery atherosclerotic plaques from e-cig smokers. Intriguingly, we found a significant increase of oxidative mitochondria DNA lesion in the plasma of ECV-exposed mice. Administration of TLR9 antagonist before ECV exposure not only alleviated atherosclerosis and the upregulation of TLR9 in plaques but also attenuated the increase of plasma levels of inflammatory cytokines, reduced the plaque accumulation of lipid and macrophages, and decreased the frequency of blood CCR2+ (C-C chemokine receptor type 2) classical monocytes. Surprisingly, we found that cytoplasmic mitochondrial DNA isolated from ECV extract-treated macrophages can enhance TLR9 activation in reporter cells and the induction of inflammatory cytokine could be suppressed by TLR9 inhibitor in macrophages.
Conclusions:
E-cig increases level of damaged mitochondrial DNA in circulating blood and induces the expression of TLR9, which elevate the expression of proinflammatory cytokines in monocyte/macrophage and consequently lead to atherosclerosis. Our results raise the possibility that intervention of TLR9 activation is a potential pharmacological target of ECV-related inflammation and cardiovascular diseases.
21. Liquid Biopsy: Current Status and Future Directions. Front Line Genomics. Jan 2021.
Case study: a new scalable and automatable method for the extraction of cfDNA.
Liquid biopsies represent a promising area of cancer testing as taking blood is less invasive than tumor biopsies. The cell free DNA (cfDNA) present in the blood includes DNA derived from cancer cells and cancer biomarkers can be detected in the extracted cfDNA. Whole blood also contains genomic DNA, and can be removed by centrifugation, resulting in plasma. cfDNA is present in very small amounts in blood or plasma, and thus larger amounts of plasma are required for many applications. Larger extractions are more challenging to automate, as they require additional pipetting steps. Here we present a novel cfDNA extraction kit and show its compatibility with extractions from 200 μl – 5 mL. We discuss the optimisation of the method and demonstrate automation on a KingFisher Duo. This workflow can also be automated on a Biomek i7 Automated Workstation. We demonstrate that this kit can be used for NGS and produce results comparable to other commercial kits.
Publications (2020)
20. Apostle COVID-19 RNA Extraction System Applied in the Effective Detection of SARS-CoV-2. Horner S. Apostle Inc. Application Note. Oct 2020.
The current coronavirus disease 2019 (COVID-19) pandemic started in late 2019. COVID-19 is the result of severe acute respiratory syndrome 2 (SARS-CoV-2) virus contraction. COVID-19 is often accompanied by a wide range of symptoms including fever, cough, and shortness of breath. The SARS-CoV-2 virus consists of a ~30 kb RNA genome encoding for 15 proteins, including the spike protein that enables the virus to enter host cells. The current gold standard qualitative detection method, qRT-PCR, reverse transcribes the viral RNA into cDNA, which is subsequently amplified and quantitated. This application note illustrates the effective detection of SARS-CoV-2 using the Apostle COVID-19 Viral RNA Isolation Automation System and qRT-PCR in clinical lab settings. This system uses efficient magnetic nanoparticle technology for fast extraction and purification of viral nucleic acids from various types of biological samples collected in transport media. The proficient and consistent systems provide reliable test results to individuals that contribute to COVID-19 pandemic relief.
The use of Apostle COVID-19 Viral RNA Isolation Automation System has proven to be an effective process as a part in detecting SARS-CoV-2 from more than two million samples carried out by our clients in clinical lab settings.
19. Liquid Biopsies Generate Ample Insights from Sparse Clues. Jon Kelvey Genetic Engineering & Biotechnology News. Vol. 40, No. 8, August 1, 2020; pp. 44–46, 48, 49
(Note: Apostle technologies are discussed in this news article.)
Duplex sequencing, target enrichment, and other technologies reveal incipient and residual disease as well early signs of drug resistance. [pdf]
18. High-resolution DNA size enrichment using a magnetic nano-platform and application in non-invasive prenatal testing. Bo Zhang, Shuting Zhao, Hao Wan, Ying Liu, Fei Zhang, Xin Guo, Wenqi Zeng, Haiyan Zhang, Linghua Zeng, Jiale Qu, Ben-Qing Wu, Xinhong Wan, Charles R. Cantor and Dongliang Ge. Analyst. July 2020, 145, 5733-5739
Precise DNA sizing can boost sequencing efficiency, reduce cost, improve data quality, and even allow sequencing of low-input samples, while current pervasive DNA sizing approaches are incapable of differentiating DNA fragments under 200 bp with high resolution (<20 bp). In non-invasive prenatal testing (NIPT), the size distribution of cell-free fetal DNA in maternal plasma (main peak at 143 bp) is significantly different from that of maternal cell-free DNA (main peak at 166 bp). The current pervasive workflow of NIPT and DNA sizing is unable to take advantage of this 20 bp difference, resulting in sample rejection, test inaccuracy, and restricted clinical utility. Here we report a simple, automatable, high-resolution DNA size enrichment workflow, named MiniEnrich, on a magnetic nano-platform to exploit this 20 bp size difference and to enrich fetal DNA fragments from maternal blood. Two types of magnetic nanoparticles were developed, with one able to filter high-molecular-weight DNA with high resolution and the other able to recover the remaining DNA fragments under the size threshold of interest with >95% yield. Using this method, the average fetal fraction was increased from 13% to 20% after the enrichment, as measured by plasma DNA sequencing. This approach provides a new tool for high-resolution DNA size enrichment under 200 bp, which may improve NIPT accuracy by rescuing rejected non-reportable clinical samples, and enable NIPT earlier in pregnancy. It also has the potential to improve non-invasive screening for fetal monogenic disorders, differentiate tumor-related DNA in liquid biopsy and find more applications in autoimmune disease diagnosis.Two types of magnetic nanoparticles were developed, with one able to filter high-molecular-weight DNA with high resolution and the other able to recover the remaining DNA fragments under the size threshold of interest with >95% yield. Using this method, the average fetal fraction was increased from 13% to 20% after the enrichment, as measured by plasma DNA sequencing. This approach provides a new tool for high-resolution DNA size enrichment under 200 bp, which may improve NIPT accuracy by rescuing rejected non-reportable clinical samples, and enable NIPT earlier in pregnancy. It also has the potential to improve non-invasive screening for fetal monogenic disorders, differentiate tumor-related DNA in liquid biopsy and find more applications in autoimmune disease diagnosis.
17. Improved conversion in extraction, library construction, and capture improve sensitivity for variants in liquid biopsy samples. Nicole Roseman, Shilpa Parakh, Hsiao-Yun Huang, Kevin Lai, Timothy Barnes, Lyn Lewis, Ushati Das Chakravarty, Anastasia Potts, Alisa Jackson, Amy Yoder, Jessica Sheu, Tzu-Chun Chen. In: Proceedings of the Annual Meeting of the American Association for Cancer Research 2020; 2020 Apr 27-28 and Jun 22-24. Philadelphia (PA): AACR; Cancer Res 2020;80(16 Suppl):Abstract nr 5863.
Demonstrate performance of a complete automation and reagent workflow for analysis of cfDNA from bodily fluids. The efficient extraction of cfDNA from bodily fluids is a unique challenge due to the very low concentrations of nucleic acid. The extraction process along with library preparation is a laborious workflow, where human variability can lead to increased variability in the downstream analysis. Integrated DNA Technology (IDT) and Beckman Coulter (BC) have teamed up to provide a complete automation and reagent workflow for analysis of low frequency variants in cfDNA. The Apostle MiniMax™ High Efficiency Isolation Kit from BC provides complex, utilized magnetic nanoparticles to effectively capture cfDNA. IDT's library prep kit utilizes novel chemistry to maximize conversion, suppress adapter-dimer formation, reduce chimera rates, and facilitate double strand consensus analysis to call ultra-low frequency variants. Finally, IDT's xGen™ hybrid capture products maintain high library diversity and on-target rates to enable low frequency variant calling regardless of panel size. The Biomek i5 and i7 Hybrid workstations bring out the best performance from these reagents. The Biomek NGS workstations protocol is written with a modular design with safe stop points, making it customizable for each lab. The automated protocol uses Beckman's Demonstrated Method Interface tools which include: Biomek Method Launcher to run the method without going into Biomek software, Method Options Selector to choose the run parameters with a user friendly interface, Guided labware Setup to set the deck with labware based on the run parameters, DeckOptix Final Check software to help reduce deck setup errors. We demonstrate the performance of this complete workflow with a range of plasma inputs (4-8 mL). Using control samples with known variant frequencies, the workflow yields high library complexity, 100% positive predictive value, and reliable detection of <0.5% mutant allele frequency variants. With real cfDNA, the workflow demonstrates both high cfDNA and sequencing library yields along with high library complexity. The combination of these reagents on the Biomek workstations provides a robust and reproducible solution for the analysis of cfDNA.
16. G3viz: an R package to interactively visualize genetic mutation data using a lollipop-diagram. Xin G, Bo Z, Wenqi Z, et al. Bioinformatics. 2020; 36(3):928–929
The lollipop-diagram is one of the widely used graphical representations to visualize and explore translational effects of genetic mutations in cancer genomics. However, an easy-to-use lollipop-diagram tool with full functionality is still lacking. Here, we introduce g3viz, an R package that enables researchers to explore genetic mutation data using a lollipop-diagram in a web browser. With a few lines of R code, users can interactively visualize data details, annotate findings and export resultant diagrams in high-quality figures. Because of usefulness and usability, g3viz can be generally exploited by researchers with different levels of bioinformatics skills and programming experience.
15. An end-to-end automated workflow for low-frequency variant detection in cfDNA. Tzu-Chun Chen, Nicole Roseman, Han Wei, et al. ASHG. Oct 27-30, 2020
Introduction
Liquid biopsy, as a tool to detect tumor-associated variants in cell-free DNA (cfDNA) from plasma, is a powerful non-invasive approach for biomarker discovery in oncology research. Integrated DNA Technologies (IDT) and Beckman Coulter Life Sciences (BECLS) have teamed up to provide a comprehensive end-to-end automated solution on the Biomek workstations for low frequency variant detection in cfDNA using next generation sequencing. The combined workflow including reagents for cfDNA extraction from plasma, NGS library preparation and hybrid capture, provides a robust and reproducible solution analyzing cfDNA samples.
Conclusions
• We provide an end-to-end automation and reagent workflow from plasma to cfDNA analysis that includes Apostle Minimax, xGenPrism DNA Library Prep Kit, and xGen hybridization capture.
• Apostle MiniMax High Efficiency Isolation Kit provides high yield extraction of cfDNA. • The xGen Prism DNA library preparation kit, optimized for low input and degraded
samples such as cfDNA, provides sensitive and accurate detection of ultra-low frequency variants.
14. A complete automation and reagent workflow for analysis of cfDNA: from plasma to variants. Roseman N, Parakh S, Lai K, Sheu J, Wei H, Niccum B, Chen T, Huang H, Barnes T, Lewis L, Chakravarty UD, and Potts A. Advances in Genome Biology and Technology (AGBT). (abstract #511). Marco Island, FL. Feb 23-26, 2020
As cost of sequencing continues to drop, the throughput and complexity of NGS assays have risen precipitously. At the same time, the types of samples being used for these assays have expanded as researchers find value in studying low input, degraded biological samples, such as cell free DNA (cfDNA). However, efficient extraction of cfDNA from bodily fluids is challenging due to the very low concentrations of nucleic acid. Manual protocols involved in the NGS workflow – DNA extraction, library preparation, and target enrichment – are often laborious and require significant hands on time, which can increase variability in the downstream analysis. Thus, complete automation workflows can increase throughput, reduce hands-on time, and minimize error. Integrated DNA Technologies (IDT) and Beckman Coulter (BC) have teamed up to provide a complete automation and reagent workflow for analysis of low frequency variants in cfDNA. The Apostle MiniMaxTM High Efficiency Isolation Kit from BC is a magnetic nanoparticle based kit that extracts cfDNA from all bodily fluids. IDT’s xGenTM Prism DNA library prep kit provides novel chemistry to maximize conversion, suppress adapter-dimer formation, reduce chimera rates, and facilitate UMI-based error correction to call ultra-low frequency variants. IDT’s xGenTM target enrichment products maintain high library diversity and on-target rates to enable low frequency variant calling regardless of panel size. The combination of these reagents on the Biomek workstations provides a robust and reproducible solution for the analysis of cfDNA.
Conclusion
• Apostle Minimax, xGen Prism, and xGen hybridization capture provide a complete automated solution from plasma to sequencing on the Beckman Coulter Biomek platforms
• Apostle Minimax provides higher quantity and quality cfDNA extraction compared to a competitor kit
• xGen Prism is specifically designed for low input, degraded samples, such as cfDNA and generates higher coverage and complexity compared to competitor kits
• UMI-based error correction can be used to eliminate nearly all false positive calls and enables accurate low frequency variant calling
13. cfDNA Extraction Efficiency Affects NGS Data. Han Wei, Ph.D., Beckman Coulter Life Sciences. Indianapolis, IN. Technical Note 2020.

Figure 1. cfDNA extraction method affects allele frequency (AF%). EGFR L858R standards with AF% of 0.3%, 1.0%, and 3.0% were spiked into plasma and isolated using Product A (red) or Apostle MiniMax cfDNA isolation kit (blue and green). The eluates were analyzed by NGS (performed by the Institute B) following standard protocol and AF% calculated using their established workflow. cfDNA isolated by Apostle MiniMax cfDNA isolation kit shows higher concordance between detected AF% and expected AF%.
In order to understand how cfDNA extraction efficiency affects NGS data, we present Institute B titration data. EGFR L858R standards with an allele frequency (AF%) of 0.3%, 1.0%, and 3.0% were spiked into plasma and isolated using either Apostle MiniMaxTM High Efficiency cfDNA Isolation or Product A. The cfDNA was analyzed via NGS and AF% was calculated by Institute B. cfDNA isolated by Apostle MiniMax cfDNA isolation kit shows concordance between detected AF% and expected AF%.
For cancer samples, the allele frequency represents the percentage of sequence reads carrying a mutant allele of an individual patient’s cancer. Researchers often set thresholds for defining the variant allele frequency as mutation; therefore, low cfDNA recovery efficiency can decrease sensitivity and lead to an increase in false negatives for mutation calling.
Publications (2019)
12. Correlation between mutations found in FFPE tumor tissue and paired cfDNA samples. (n=8) Niccum B., Saunders L., Hur A.,Patel A. The American Society of Human Genetics (ASHG). (abstract #1766). Houston, TX. Oct 15, 2019
Liquid biopsies represent a promising area of facilitating cancer research as taking blood is less invasive than tumor biopsies. Cell-free DNA (cfDNA) consists of small (150 – 500 bp) DNA fragments that circulate in the blood. Levels of cfDNA tend to be low in healthy, non-pregnant patients and increased in patients with cancer, pregnancy, or extensive tissue damage. cfDNA is believed to be derived mostly from apoptotic cells and a source for biomarkers for a variety of diseases.
As a non-invasive way to detect disease cfDNA is extracted from blood; however, there is some concern that cfDNA does not contain the same biomarkers as tumor tissue. Tumor tissue is typically removed and stored as formalin-fixed, paraffin-embedded tissue, a process that preserves the morphological structures well but chemically modifies and degrades the nucleic acids.
Despite the difficulties, FFPE tissue (shown on the left) is often used to look for cancer-associated mutations; however it does not always correlate with the mutations seen in cfDNA. In this poster we present a comparison of matched FFPE and plasma samples to determine how many mutation are seen in both tissues. We also look at where the mutational mismatches appear in the chromosome. We found chromosomal regions have different mismatch rates, and we use this to draw conclusions about the best chromosomal locations for biomarkers. We also look at the different results that can come from using multiple different panels.
Here we show that sequencing of cfDNA captures the majority of variants that are found when sequencing FFPE DNA. One result shown here is that more indels are identified using cfDNA than with FFPE DNA. This is especially shown here with breast tissue, while this is inconclusive with lung tissue. More samples should be tested before any conclusions can be derived.
Another interesting finding is the distribution of variants across the genome. There is some indication that variants sequenced using cfDNA is correlated better to the variants found with either cfDNA or FFPE. This result could be used to better understand if there is bias that occurs when sequencing FFPE DNA and where this bias could be from such as cross-linking could be more apparent in parts of chromosomes.
11. Comparison between mutation profiles of paired whole blood and cfDNA samples. Patel A., Saunders L., Hur A. The American Society of Human Genetics (ASHG). (abstract #1767). Houston, TX. Oct 15, 2019
Liquid biopsies are increasingly becoming a tool of choice for researching cancer detection and monitoring. Cell-free DNA or cfDNA is simply small fragments of DNA circulating in bodily fluids. It is also known as circulating cell-free DNA (ccfDNA), circulating tumor DNA (ctDNA) and cell free-fetal DNA (cffDNA). Next-Gen Sequencing of cfDNA is coming into maturity as a non-invasive method to identify mutational profiles in many cancer types.
A big question is how do you separate a germ line variant from a tumor variant. Understanding the difference in a patient sample can give a more thorough understanding of a variant that can be used to study a cancer type. An easy solution is to compare germ line variants from whole blood genomic DNA (gDNA). This would couple easily with plasma sample collection as plasma can be directly separated from a single sample point.
Here we describe a simple method to isolate both gDNA and cfDNA from a donor blood sample and discuss the automation of both extractions. We show the efficacy of cfDNA as reliable biomarker analysis tool by comparing mutations in cfDNA vs whole blood. The study determines if the difference between tumor and germ line mutations can be established and the limitations.
Due to larger volumes necessary to extract sufficient concentrations of cfDNA, automation can assist in the extraction. The Apostle MiniMax™ High Efficiency Cell-Free DNA (cfDNA) extraction kit was automated on the Biomek i-Series. It provides equal recovery of cfDNA as a manual extraction with much less hands on-time. The kit used in the study to extract whole blood, GenFind V3, has also been automated on the Biomek i-Series; allowing for reduced hands with the same quality results as a manual extraction.
Conclusions
• Variants found only in cfDNA could be used as an initial screen for ctDNA analysis
• Apostle MiniMax™ and GenFind V3 can be used together to get a picture of germ line variants and cfDNA, potential ctDNA, variants
• These results show that a holistic view of a cancer subject can be gained by using one sample source, whole blood
10. Correlation between mutations found in FFPE tumor tissue and paired cfDNA samples. Niccum B., Heath C., Saunders L., Hur A.,Patel A. Association for Molecular Pathology (AMP). (abstract #ST103). Baltimore, MD. November 7-9, 2019
Liquid biopsies represent a promising area of facilitating cancer research as blood collection is less invasive than tumor biopsies. Cell free DNA (cfDNA) consists of small (150 – 500 bp) DNA fragments that circulate in the blood. cfDNA levels tend to be low in healthy, non-pregnant patients, and increase in patients with cancer, pregnancy, or extensive damage to tissue. cfDNA is believed to be derive mostly from apoptotic cells for which biomarkers for a variety of diseases have been found in cfDNA.
FFPE tissue is often used to look for cancer-associated mutations despite invasiveness; however it does not always correlate with the mutations seen in cfDNA. In this poster we present a comparison of matched FFPE and plasma samples to determine how many mutation are seen in both tissues. We also look at where the mutational mismatches appear in the chromosome. Different chromosomal regions can have different mismatch rates, and we use this to draw conclusions about the best chromosomal locations for biomarkers. We automated from extraction through sequencing in collaboration with Swift biosciences.
As cfDNA is extracted from blood, it is a non-invasive way to detect disease; however, there is some concern that cfDNA does not contain the same biomarkers as tumor tissue. Tumor tissue is typically removed and stored as formalin-fixed, paraffinembedded tissue, a process that preserves the morphological structures well but chemically modifies and degrades the nucleic acids.
Here we show:
• Sequencing of cfDNA captures the majority of variants that found in sequenced FFPE DNA
• More indels are identified using cfDNA than with FFPE DNA, especially with breast tissue
• Distribution of variants across the genome differs when sequencing FFPE DNA
• More previously identified clinically relevant variants, as identified by the ClinVar database were found when sequencing FFPE DNA
This study is small and further work should be done using larger data sets to gain more conclusive information.
9. Dynamics of Plasma EGFR T790M Mutation in Advanced NSCLC: A Multicenter Study. Yang et al. Targeted Oncology. 2019;14:719-728. Published: 06 November 2019.
(Note: Apostle MiniMax technology is used in this clinical study.)
Background Droplet digital polymerase chain reaction (ddPCR) is an emerging technology for quantitative cell-free DNA oncology applications. However, a ddPCR assay for the epidermal growth factor receptor (EGFR) p.Thr790Met (T790M) mutation suitable for clinical use remains to be established with analytical and clinical validations. Objective We aimed to develop and validate a new ddPCR assay to quantify the T790M mutation in plasma for monitoring and predicting the progression of advanced non-small-cell lung cancer (NSCLC). Methods Specificity of the ddPCR assay was evaluated with genomic DNA samples from healthy individuals. The inter- and intraday variations of the assay were evaluated using mixtures of plasmid DNA containing wild-type EGFR and T790M mutation sequences. We assessed the clinical utility of the T790M assay in a multicenter prospective study in patients with advanced NSCLC receiving tyrosine kinase inhibitor (TKI) treatment by analyzing longitudinal plasma DNA samples. Results We set the criteria for a positive call when the following conditions were satisfied: (1) T790M mutation frequency > 0.098% (3 standard deviations above the background signal); (2) at least two positive droplets in duplicate ddPCR reactions. Among the 62 patients with advanced NSCLC exhibiting resistance to TKI treatment, 15 had one or more serial plasma samples that tested positive for T790M. T790M mutation was detected in the plasma as early as 205 days (median 95 days) before disease progression, determined by imaging analysis. Plasma T790M concentrations also correlated with intervention after disease progression. Conclusions We developed a ddPCR assay to quantify the T790M mutation in plasma. Quantification of longitudinal plasma T790M mutation may allow noninvasive assessment of drug resistance and guide follow-up treatment in TKI-treated patients with NSCLC. Trial Registration Clinical Trials.gov identifier: NCT02804100.
8. Isolation of cell-free DNA (cfDNA) from plasma using Apostle MiniMaxTM High Efficiency cfDNA Isolation kit—comparison of fully automated, semi-automated and manual workflow processing. Brittany Niccum, PhD., Randy Pares and Antonia Hur. Beckman Coulter Life Sciences. Application Note. Sept 2019.
Cell-free DNA is present in plasma, urine, and other bodily fluids. Typically cfDNA is at low concentration and comprises double-stranded DNA fragments that are overwhelmingly short (140 to 180 base pairs). Its small size and low quantities present a challenge for sequencing and other downstream applications due to insufficient cfDNA yield. Subsequently, resulting in a need to extract from more substantial volumes of bodily fluid; yet higher input volumes can be more challenging to manage for high-throughput sample processing.
This application note compares workflows and yield for the extraction of cfDNA using Apostle MiniMax High Efficiency cfDNA Isolation Kit, following the manual protocol, automating the extraction using the Biomek i7 Hybrid Workstation, and semi-automating the extraction using the KingFisher Duo Prime Sample Purification System. These potential solutions help mitigate some of the challenges of processing large volume samples. Automating the chemistry can also reduce the risk of human error and reduce hands-on time, therefore giving the user the ability to run more samples in a day.
7. Cell-Free DNA Isolation Kit. Science. 17 May 2019:Vol. 364, Issue 6441, pp. 696. DOI: 10.1126/science.364.6441.696-a. (Featured in New Products section)
Surpassing industry standards for yield and purity of cell-free DNA (cfDNA), Apostle MiniMax from Beckman Coulter is a magnetic nanoparticle–based kit that extracts cfDNA from plasma using manual or automated workflows. Due to limited cfDNA concentration, sufficient yield for NGS or other downstream applications can require higher volumes of plasma. As the volume increases, issues such as recovery efficiency and workflow complexity often arise. Plasma contains a host of contaminants that are problematic at this scale and can reduce assay sensitivity. Apostle MiniMax technology performs reliably across a range of volume inputs, consistently recovering high quantities of cfDNA while effectively removing contaminants.
6. A workflow for medium-throughput isolation of cfDNA from plasma samples using Apostle MiniMaxTM on the KingFisherTM Technology. Brittany Niccum, PhD. Beckman Coulter Life Sciences. Application Note. 2019.
Cell-free DNA (cfDNA) is found in various bodily fluids. cfDNA has a characteristic size of approximately 175 bp. Due to its small size and low quantity the main challenge is to get enough cfDNA for sequencing. Subsequently, this results in a need to extract from larger volumes of bodily fluid, yet higher input volumes can be more difficult to manage for high-throughput sample processing.
This application note demonstrates the use of Apostle MiniMax High Efficiency cfDNA Isolation Kit, in conjunction with the KingFisher Duo Prime automated protein purification system. This potential solution that mitigates some of the challenges of processing large volume samples. Automating the chemistry can also reduce the risk of human error, reduce hands-on time and total time; therefore giving the user the ability to run more samples in a day.
5. Biomek Automated Genomic Sample Prep Accelerates Research: Biomek i-Series Automation of the Apostle MiniMaxTM High Efficiency cfDNA Isolation Kit. Application Overview. Beckman Coulter Life Sciences. 2019
The Apostle MiniMax High Efficiency cfDNA Isolation Kit isolates cell free DNA (cfDNA) and circulating tumor DNA (ctDNA) from plasma collected from blood collection tubes containing EDTA and other blood collection tube types as well as from serum and urine. The kit produces high quality extracted cfDNA that can be used in downstream genomic assays, such as PCR amplification and next-generation sequencing. Proteins in cell free plasma are digested and cfDNA is captured using Apostle’s proprietary technology. Contaminants are removed from the samples through several simple washes, leaving high quality cfDNA samples that are ready for elution. In this technical note, we demonstrate the automated performance of the Apostle MiniMax High Efficiency cfDNA Isolation Kit on the Biomek i7 Hybrid Genomics Workstation.
When compared to manual operations, the Apostle MiniMax High Efficiency cfDNA Isolation Kit automated on Biomek platforms provide:
• Reduced hands-on-time and increased throughput
• Option to run the method end-to-end with only setup and tear-down touch points
• Reduction in pipetting errors
• Standardized workflow for improved results
• Quick implementation with demonstrated methods
• Knowledgeable support for reagents, automation and methods all from a single vendor
4. Correlation between mutations found in FFPE tumor tissue and paired cfDNA samples. (n=3) Saunders L and Patel A. American Association for Cancer Research (AACR). (abstract #2233). Atlanta, GA. April 2, 2019
Liquid biopsies represent a promising area of facilitating cancer research as taking blood is less invasive than tumor biopsies. The cell free DNA (cfDNA) present in the blood includes DNA derived from cancer cells and cancer biomarkers can be detected in the extracted cfDNA. However, cfDNA is a less direct view of what is happening in the tumor, and can have a different genetic profile than the tumor tissue itself.
Tumor tissue is typically removed and stored as formalin-fixed, paraffin-embedded tissue, a process that preserves the morphological structures well but chemically modifies and degrades the nucleic acids. This tissue is often used to look for cancer- associated mutations despite these difficulties; however, it does not always correlate with the mutations seen in cfDNA.
In this poster we present a comparison of matched FFPE and plasma samples to determine how many mutations are seen in both tissues. We also look at where the mutational mismatches appear in the chromosome. Different chromosomal regions can have different mismatch rates, and we use this to draw conclusions about the best chromosomal locations for biomarkers.
Conclusions:
Both FFPE and cfDNA detect at least two thirds of the observed indels and at least 90% of the observed SNVs. As SNVs are more likely to be found in both tissue types, they are more suitable for biomarker use if looking across different tissues. This was true for all three cancers tested.
Different regions of the chromosome have different rates of mismatches in mutation detection between plasma and FFPE tissue. The first 10-30%, 40-50%, 60-80%, and 90-100% of the chromosome have the lowest rates of mismatch and provide the best locations for biomarkers detectible in both tissues. Future work will focus in increasing the sample size to further narrow down the areas of the chromosome with the highest likelihood of good mutation detection in both plasma and FFPE.
3. A new Scalable and automatable method for the extration of cfDNA. Saunders LP, Hur A, Niccum B, and Patel A. Advances in Genome Biology and Technology (AGBT). (abstract #419). Marco Island, FL. Feb 28, 2019
Liquid biopsies represent a promising area of cancer testing as taking blood is less invasive than tumor biopsies. The cell free DNA (cfDNA) present in the blood includes DNA derived from cancer cells and cancer biomarkers can be detected in the extracted cfDNA.
Whole blood also contains genomic DNA, and can be removed by centrifugation, resulting in plasma. cfDNA is present in very small amounts in blood or plasma, and thus larger amounts of plasma are required for many applications. Larger extractions are more challenging to automate, as they require additional pipetting steps.
Here we present a novel cfDNA extraction kit and show its compatibility with extractions from 200 μl – 5 mL. We discuss the optimization of the method and demonstrate automation on a KingFisher Duo. This workflow can also be automated on a Biomek i7 Automated Workstation. We demonstrate that this kit can be used for NGS and produce results comparable to other commercial kits.
Conclusions:
• DNA can be extracted from 200 μL to 5 mL of plasma
• The Apostle MiniMax kit removes the PCR inhibitors present in plasma
• Genomic contamination is not present in the extracted cfDNA
• Extraction of 1 mL plasma can be automated on a KingFisher instrument with yields similar to manual extraction
• Similar numbers of mutations were found in cancer plasma with the Apostle MiniMax kit and another commercial kit.
The Apostle MiniMax kit is a versatile new cfDNA kit that can extract from a wide range of sample amounts and be run either manually or on a variety of automation systems.
2. cfDNA Extraction from Plasma for Liquid Biopsy: Apostle MiniMaxTM High Efficiency cfDNA Isolation Kit. Beckman Coulter Life Sciences, Data Sheet. 2019.

(Figure 1) "For each tube the Apostle MiniMax extracted higher total yield of DNA. "
Apostle MiniMax High Efficiency cfDNA Isolation Kit, Apostle MiniMax is a cell–free DNA (cfDNA) isolation reagent kit, built on magnetic bead–based technology. Apostle MiniMax has been demonstrated to purify cfDNA from human plasma in both manual and automated workflows.
• Data representative of results of cfDNA extracted from 1–5mL of plasma
• Demonstrated compatibility with a variety of collection tubes
• cfDNA purity shown to be suitable for downstream PCR based assays
Publications (2018)
1. Competitive evolution of NSCLC tumor clones and the drug resistance mechanism of first-generation EGFR-TKIs in Chinese NSCLC patients. Deng et al. Heliyon. VOLUME 4, ISSUE 12, E01031, DECEMBER 01, 2018
Purpose - Although many studies have reported on the resistance mechanism of first-generation EGFR TKIs (1st EGFR TKIs) treatment, large-scale dynamic ctDNA mutation analysis based on liquid biopsy for non-small cell lung cancer (NSCLC) in the Chinese population is rare. Using in-depth integration and analysis of ctDNA genomic mutation data and clinical data at multiple time points during the treatment of 53 NSCLC patients, we described the resistance mechanisms of 1st EGFR TKIs treatment more comprehensively and dynamically. The resulting profile of the polyclonal competitive evolution of the tumor provides some new insights into the precise treatment of NSCLC.
Experimental design - A prospective study was conducted in patients with advanced NSCLC with acquired resistance to erlotinib, gefitinib or icotinib. By liquid biopsy, we detected mutations in 124 tumor-associated genes in the context of drug resistance. These 124 genes covered all tumor therapeutic targets and related biological pathways. During the entire course of treatment, the interval between two liquid biopsies was two months.
Results - Unlike the common mutations tested in tissue samples, our data showed a higher coverage of tumor heterogeneity (32.65%), more complex patterns of resistance and some new resistance mutation sites, such as EGFR p.V769M and KRAS p.A11V. The major resistance-associated mutations detected were still EGFR p.T790M (45.28%), other point mutations in EGFR (33.9%), and KRAS and NRAS mutations (15.09%). These mutation ratios might be considered as a preliminary summary of the characteristics of Chinese patients. In addition, starting from the two baseline mutations of the EGFR gene (19del vs. L858R), we first described the detailed mutation profile of the EGFR gene. Although there was no significant difference in the number of patients with EGFR p.19del and EGFR p.L858R baseline mutations (24% vs. 16%, P = 0.15), patients from the EGFR p.19del baseline group were much more likely to develop EGFR p.T790M resistance mutations (62.1% vs. 19.3%, P = 0.007). Through careful integration of gene mutation information and clinical phenotype information, an interesting phenomenon was found. Although the variant allele fraction (VAF) of the EGFR p.T790M mutation was significantly linearly correlated with that of the EGFR drug-sensitive mutation (r = 0.68, P = 0.00025), neither VAF was associated with the tumor volume at the advanced stage. It was shown that other tumor clones might contribute more to the resistance to 1st EGFR TKIs treatment than tumor clones carrying the EGFR p.T790M mutation when resistance developed. By further analysis, we found that, in some patients, when the primary tumor clones detected were those carrying EGFR−/− mutations (both types the EGFR p.19del/p.L858R and EGFR p.T790M mutation types were missing), most of them showed a poor prognosis and ineffective late treatment, indicating that EGFR−/− played a more important role than EGFR p.T790M in the process of NSCLC drug resistance in these patients. From the perspective of the clonal evolution of NSCLC tumor cells, these phenomena could be explained by the competitive evolution between different tumor clones. In addition, two new mutations, KRAS p.A11V and EGFR p.V769M, emerged significantly during drug resistance in NSCLC patients and had shown obvious competitive clonal evolution characteristics. Combined with clear clinical drug resistance phenotypic information, we believed that these two new mutations might be related to new drug resistance mechanisms and deserve further study. We have also seen an interesting phenomenon. In some patients undergoing 1st EGFR TKIs treatment, the EGFR p.T790M mutation appeared, disappeared, and reappeared, and this spatial and temporal diversity of the EGFR p.T790M mutation was regulated by targeted drug and chemotherapy and was correlated with the individual tumor mutation profile.
Conclusions - The constitution and competitive evolution of the tumor clones have a decisive influence on treatment and can be regulated by targeted drugs and chemotherapy. Additionally, EGFR p.T790M spatial and temporal diversity during treatment warrants more attention, and this spatial and temporal diversity may be useful for the choice of treatment strategies for certain NSCLC patients. Through longitudinal cfDNA sample analysis, the resistance mechanism and dynamic clinical features of Chinese NSCLC patients are systematically established as reliable and meaningful to understand acquired resistance and make further personalized treatment decisions dynamically. Two new potential drug resistance-associated mutations in EGFR and KRAS have been found and are worthy of further study. Finally, our research shows that the evolutionary process of tumor cloning can be artificially regulated and intervened, possibly providing a new way to treat tumors.
(Methods section) - Cell free DNA was isolated using Apostle MiniMaxTM High Efficiency cfDNA Isolation Kit (Standard Edition) according to the manufacturer's protocol.
Apostle technologies have been cited or used by scientists from the following organizations. Click to read more when there is a publication.
D. Ge
- Ge D*, Fellay J*, Thompson AJ*, Simon JS*, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009:461, 399-401.
• Featured in Nature: news & views: Genomics: Hepatitis C virus gets personal. Nature 2009; 461 (357-358).
• Nature: Outlook: Pharmacogenomics: playing the odds. Nature. 2011;474(7350):S9-10.
• Licensed to: LabCorp Inc.; Quest Diagnostics Inc.
• Citations: 3900+
• Ranked No. 7 across all scientific areas published by Nature (Google Scholar Metrics, 2015) - Fellay J*, Thompson AJ*, Ge D*, Gumbs CE, Urban TJ, Shianna KV, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464(7287):405-8. (* Equal co-authors)
- Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'hUigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 2009; 461, 798-801.
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Bottcher Y, Olesen J, Breuer R, Moller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Rethelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Group, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Lim Yoon J, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA, Kahn RS, Linszen DH, van Os J, Wiersma D, Bruggeman R, Cahn W, de Haan L, Krabbendam L, Myin-Germeys I. Common variants conferring risk of schizophrenia. Nature 2009:460(7256):744-7
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D , Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Kahn RS, Linszen D, van Os J, Wiersma D, Bruggeman R, Cahn W, Germeys I, de Haan L, Krabbendam L, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature . 2008;455(7210):232-6.
- Fellay J, Shianna KV *, Ge D *, Colombo S *, Ledergerber B *, Weale M *, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. A whole-genome association study of major determinants for host control of HIV-1. Science . 2007;317(5840):944-7. (* Equal authors) (Citations: 390)
- Thomas R, Apps R, Qi Y, Gao X, Male V, O'HUigin C, O'Connor G, Ge D, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41(12):1290-4.
- Ge D, Ruzzo EK, Shianna KV, He M, Pelak K, Heinzen EL, Need AC, Cirulli ET, Maia JM, Zhu M, Singh A, Allen AS, Goldstein DB. SVA: Software for Annotating and Visualizing Sequenced Human Genomes. Bioinformatics, 2011;27(14):1998-2000. (software: http://www.svaproject.org )
- Zhu Q *, Ge D*, Maia JM, Zhu M, Petrovski S, Dickson SP, Heinzen EL, Shianna KV, Goldstein DB. A Genome-Wide Comparison of the Functional Properties of Rare and Common Genetic Variants in Humans. American Journal of Human Genetics. 2011;88(4):458-68.
- W. J. Sandborn, B. R. Bhandari, R. Fogel, J. Onken, E. Yen, X. Zhao, Z. Jiang, D. Ge, Y. Xin, Z. Ye, D. French, J. A. Silverman, B. Kanwar, G. M. Subramanian, J. G. McHutchison, S. D. Lee, L. M. Shackelton, R. K. Pai, B. G. Levesque & B. G. Feagan. Randomised clinical trial: a phase 1, dose-ranging study of the anti-matrix metalloproteinase-9 monoclonal antibody GS-5745 versus placebo for ulcerative colitis. Alimentary Pharmacology and Therapeutics. 2016 (In press)
- Charuworn P, Hengen PN, Aguilar Schall R, Dinh P, Ge D, Corsa A, Reesink HW, Zoulim F, Kitrinos KM. Baseline interpatient hepatitis B viral diversity differentiates HBsAg outcomes in patients treated with tenofovir disoproxil fumarate. J Hepatol. 2015 May;62(5):1033-9.
- Nelson D, Yoshida EM, Paulson MS, Hengen MS, Hengen PN, Ge D, Kanwar B, McNally J, Pang PS, Subramanian GM, McHutchison JG, Urbanek P, Lawitz E, Urban TJ. Genome-wide association study to characterize serum bilirubin elevations in patients with HCV treated with GS-9256, an HCV NS3 serine protease inhibitor. Antiviral Therapy. 2014 Feb 6. doi: 10.3851/IMP2747.
- Pelak K*, Shianna KV*, Ge D*, Maia JM, Zhu M, Smith JP, et al. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6(9): e1001111.
- Sobreira NLM, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, Ge D, et al. Whole-Genome Sequencing of a Single Proband Together with Linkage Analysis Identifies a Mendelian Disease Gene. PLoS Genet 2010;6(6):e1000991.
- Todd J, Goldstein DB, Ge D, Christie J, Palmer, SM. The State of Genome-Wide Association Studies in Pulmonary Disease: A New Perspective. American Journal of Respiratory and Critical Care Medicine. 2011.
- Thompson AJ, Muir AJ, Sulkowski MS, Patel K, Tillmann HL, Clark PJ, Naggie S, Fellay J, Ge D, McCarthy JJ et al: Hepatitis C trials that combine investigational agents with pegylated interferon should be stratified by interleukin-28B genotype. Hepatology 2010, 52(6):2243-2244.
- Thompson AJ, Clark PJ, Singh A, Ge D, et al. Genome-wide association study of interferon-related cytopenia in chronic hepatitis C patients. J Hepatol. 2011.
- Thompson AJ, Fellay J, Patel K, Tillmann HL, Naggie S, Ge D, et al. Variants in the ITPA Gene Protect Against Ribavirin-Induced Hemolytic Anemia and Decrease the Need for Ribavirin Dose Reduction. Gastroenterology. 2010;139(4):1181-9.
- Thompson AJ, Muir AJ, Sulkowski MS, Ge D, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139(1):120-9 e18.
- Walley NM, Julg B, Dickson SP, Fellay J, Ge D, Walker BD, Carrington M, Cohen MS, de Bakker PI, Goldstein DB, Shianna KV, Haynes BF, Letvin NL, McMichael AJ, Michael NL, Weintrob AC. The Duffy antigen receptor for chemokines null promoter variant does not influence HIV-1 acquisition or disease progression. Cell Host Microbe 2009; 5 (5) : 408-10.
- Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KL, Hunt P, Ge D, Heinzen EL, Maia JM, Shianna KV, Weale ME, Cherkas LF, Clement G, Spector TD, Gibson G, Goldstein DB. A Genome-wide Study of Common SNPs and CNVs in Cognitive Performance in the CANTAB battery. Hum Mol Genet 2009.
- Pillai SG, Ge D *, Zhu G*, Kong X*, Shianna KV, Need AC, S. F, Hersh CP, Bakkgators, Rennard SI, Lomas D, Silverman EK, Goldstein DB. A Genome-wide Association Study in Chronic Obstructive Pulmonary Disease (COPD): Identification of two Major Susceptibility Loci. PLoS Genet . 2009; 5(3): e1000421. doi:10.1371/journal.pgen.1000421 (* Equal authors)
- Need AC *, Ge D * , Maia J, Shianna KV, Feng S, Strittmatter WJ, McEvoy JP, Keefe RSE, St Jean PL, Giegling I, Hartmann AM, M?ller H, Ruppert A, Fraser G, Crombie C, Francks C, St.Clair D, Roses AD, Muglia P, Rujescu D, Goldstein DB. A Genome-Wide Investigation of SNPs and CNVs in Schizophrenia. PLoS Genet. 2009; 5(2):e1000373.(* Equal authors)
- Heinzen E *, Ge D *, Cronin KD, Maia J, Shianna KV, Gabriel W, Welsh-Bohmer KA, Hulette CM, Denny T, Goldstein DB. Tissue specific genetic control of gene expression and alternative splicing: Implications for the study of human complex traits. PLoS Biol . 2008; 6(12): e1000001. (* Equal authors)
- Ge D , Zhang K, Need AC, Martin O, Fellay J, Urban TJ, Telenti A, Goldstein DB. WGAViewer: Software for genomic annotation of whole genome association studies. Genome Research. 2008;18(4):640-3. (citations: 87)
- Price AL, Weale ME, Patterson N, Myers SR, Need AC, Shianna KV, Ge D , Rotter JI, Torres E, Taylor KD, Goldstein DB, Reich D. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet 2008 . 83(1):132-5.
- Cavalleri GL, Weale ME, Shianna KV, Singh R, Lynch JM, Grinton B, Szoeke C, Murphy K, Kinirons P, O'Rourke D, Ge D , Depondt C, Claeys KG, Pandolfo M, Gumbs C, Walley N, McNamara J, Mulley JC, Linney KN, Sheffield LJ, Radtke RA, Tate SK, Chissoe SL, Gibson RA, Hosford D, Stanton A, Graves TD, Hanna MG, Eriksson K, Kantanen AM, Kalviainen R, O'Brien TJ, Sander JW, Duncan JS, Scheffer IE, Berkovic SF, Wood NW, Doherty CP, Delanty N, Sisodiya SM, Goldstein DB. Multicentre search for genetic susceptibility loci in sporadic epilepsy syndrome and seizure types: a case-control study. Lancet Neurol. 2007;6(11):970-80.
- Valdes AM, Loughlin J, Timms KM, van Meurs JJ, Southam L, Wilson SG, Doherty S, Lories RJ, Luyten FP, Gutin A, Abkevich V, Ge D, Hofman A, Uitterlinden AG, Hart DJ, Zhang F, Zhai G, Egli RJ, Doherty M, Lanchbury J, Spector TD. Genome-wide association scan identifies a prostaglandin-endoperoxide synthase 2 variant involved in risk of knee osteoarthritis. Am J Hum Genet 2008;82(6):1231-40.
- Cronin KD, Ge D, Manninger P, Linnertz C, Rossoshek A, Orrison BM, Bernard DJ, El-Agnaf OM, Schlossmacher MG, Nussbaum RL, Chiba-Falek O. Expansion of the Parkinson Disease-Associated SNCA-Rep1 Allele Up-Regulates Human {alpha}-Synuclein in Transgenic Mouse Brain. Hum Mol Genet 2009.
- Need AC, Keefe RS, Ge D, Grossman I, Dickson S, McEvoy JP, Goldstein DB. Pharmacogenetics of antipsychotic response in the CATIE trial: a candidate gene analysis. Eur J Hum Genet 2009; 17 (7) : 946-57 .
- Ge D , Su S, Zhu H, Dong Y, Wang X, Harshfield GA, Treiber FA, Snieder H. Stress-Induced Sodium Excretion. A New Intermediate Phenotype to Study the Early Genetic Etiology of Hypertension? Hypertension 2009;53:262-269
- Ge D , Gooljar SB, Kyriakou T, Collins LJ, Swaminathan R, Snieder H, Spector TD, O'Dell SD. Association of Common JAK2 Variants With Body Fat, Insulin Sensitivity and Lipid Profile. Obesity (Silver Spring) 2008;16(2) : 492-6.
- Ge D, Zhu H, Huang Y, Treiber FA, Harshfield GA, Snieder H, Dong Y. Multilocus analyses of Renin-Angiotensin-aldosterone system gene variants on blood pressure at rest and during behavioral stress in young normotensive subjects. Hypertension . 2007 ;49(1):107-12.
- Ge D, Young TW, Wang X, Kapuku GK, Treiber FA, Snieder H. Heritability of arterial stiffness in black and white American youth and young adults. Am J Hypertens 2007;20(10) : 1065-72.
- Kapuku GK, Ge D, Vemulapalli S, Harshfield GA, Treiber FA, Snieder H. Change of genetic determinants of left ventricular structure in adolescence: longitudinal evidence from the Georgia cardiovascular twin study. Am J Hypertens 2008;21(7):799-805.
- Oberg S, Ge D, Cnattingius S, Svensson A, Treiber FA, Snieder H, Iliadou A. Ethnic differences in the association of birth weight and blood pressure the georgia cardiovascular twin study. Am J Hypertens 2007;20(12):1235-41.
- Zhu H, Yan W, Ge D , Treiber FA, Harshfield GA, Kapuku G, Snieder H, Dong Y. Relationships of cardiovascular phenotypes with healthy weight, at risk of overweight, and overweight in US youths. Pediatrics 2008;121(1) : 115-22.
- Wang L, Li B, Lu X, Zhao Q, Li Y, Ge D, Li H, Zhang P, Chen S, Chen R, Qiang B, Gu D. A functional intronic variant in tyrosine hydroxylase (TH) gene confers risk of essential hypertension in northern Chinese Han population. Clin Sci (Lond) 2008.
- Dalageorgou C, Ge D , Jamshidi Y, Nolte IM, Riese H, Savelieva I, Carter ND, Spector TD, Snieder H. Heritability of QT Interval: How Much Is Explained by Genes for Resting Heart Rate? J Cardiovasc Electrophysiol 2007.
- Morell RJ, Brewer CC, Ge D , Snieder H, Zalewski CK, King KA, Drayna D, Friedman TB. A twin study of auditory processing indicates that dichotic listening ability is a strongly heritable trait. Hum Genet 2007;122(1):103-11.
- Zhu H, Yan W, Ge D, Treiber FA, Harshfield GA, Kapuku G, Snieder H, Dong Y. Cardiovascular characteristics in American youth with prehypertension. Am J Hypertens 2007;20(10):1051-7.
- Jamshidi Y, Snieder H, Ge D , Spector TD, O'Dell SD. The SH2B gene is associated with serum leptin and body fat in normal female twins. Obesity (Silver Spring) . 2007;15(1):5-9.
- Healey PR, Mitchell P, Gilbert CE, Lee AJ, Ge D , Snieder H, Spector TD, Hammond CJ. The inheritance of peripapillary atrophy. Invest Ophthalmol Vis Sci 2007;48(6):2529-34.
- Weili Y, He B, Yao H, Dai J, Cui J, Ge D , Zheng Y, Li L, Guo Y, Xiao K, Fu X, Ma D. Waist-to-height ratio is an accurate and easier index for evaluating obesity in children and adolescents. Obesity (Silver Spring) 2007;15(3):748-52.
- Jamshidi Y, Gooljar SB, Snieder H, Wang X, Ge D , Swaminathan R, Spector TD, O'Dell SD. SHP-2 and PI3-kinase genes PTPN11 and PIK3R1 may influence serum apoB and LDL cholesterol levels in normal women. Atherosclerosis 2007; 194 (2) : e26-33
- Ge D, Dong Y, Wang X, Treiber FA, Snieder H. The Georgia Cardiovascular Twin Study: influence of genetic predisposition and chronic stress on risk for cardiovascular disease and type 2 diabetes. Twin Res Hum Genet . 2006;9(6):965-70.
- Spencer-Jones NJ*, Ge D *, Snieder H, Perks U, Swaminathan R, Spector TD, Carter ND, O'Dell SD. AMP-kinase alpha2 subunit gene PRKAA2 variants are associated with total cholesterol, low-density lipoprotein-cholesterol and high-density lipoprotein-cholesterol in normal women. Journal of Medical Genetics. 2006 ;43(12):936-42. (* Joint authors.)
- Gu D, Ge D, Snieder H, He J, Chen S, Huang J, Li B, Chen R, Qiang B. Association of alpha-1A adrenergic receptor gene variants on chromosome 8p21 with human stage-2 hypertension. Journal of Hypertension , 2006;24(6):1057-1064.
- Kupper N, Ge D, Treiber FA, Snieder H . Tracking of blood pressure and underlying hemodynamics in European and African American adolescents. Stable heritabilities and expression of new genes. Hypertension , 2006 47(5):948-54 .
- Gu D, Su S, Ge D, Chen S, Huang J, Li B, Chen R, Qiang B. An Association Study with 33 SNPs in 11 Candidate Genes for Hypertension in Chinese. Hypertension , 2006;47(6):1147-54.
- Herold SE, Young TW, Ge D, Snieder H, Lovrekovic GZ. Sleep Disordered Breathing in Pediatric Patients with Tetralogy of Fallot. Pediatric Cardiology , 2006;27(2):243-9.
- de Lange M, Andrew T, Snieder H, Ge D, Futers TS, Standeven K, Spector TD, Grant PJ and Ariens RAS. Joint linkage and association of 6 single nucleotide polymorphisms in the factor XIII-A subunit gene point to V34L as the main functional locus. Arteriosclerosis, Thrombosis, and Vascular Biology . 2006 Aug;26(8):1914-9.
- Yang W, Huang J, Yao C, Su S, Liu D, Ge D , Gu D. Linkage and linkage disequilibrium analysis of the lipoprotein lipase gene with lipid profiles in Chinese hypertensive families. Clinical Science (Lond). 2005;108(2):137-142.
- Ge D, Huang J, Yang W, Zhao J, Shen Y, Qiang B, Gu D. Linkage analysis of chromosome 1 with essential hypertension and blood pressure quantitative traits in Chinese families. Annals of Human Genetics . 2005;69(Pt 1):45-54.
- Ge D , Huang J, He J, Li B, Duan X, Chen R, Gu D. beta2-Adrenergic receptor gene variations associated with stage-2 hypertension in northern Han Chinese. Annals of Human Genetics . 2005;69(Pt 1):36-44.
- Li B, Ge D , Wang Y, Zhao W, Zhou X, Gu D, Chen R. G Protein beta3 Subunit Gene Variants and Essential Hypertension in the Northern Chinese Han Population. Annals of Human Genetics . 2005;69(Pt 4):468-473.
- Chen S, Yan W, Huang J, Ge D, Yao Z, Gu D. Association analysis of the variant in the regulatory subunit of phosphoinositide 3-kinase (p85alpha) with Type 2 diabetes mellitus and hypertension in the Chinese Han population. Diabetic Medicine . 2005;22(6):737-743.
- Yan W, Yang X, Zheng Y, Ge D , Zhang Y, Shan Z , Simu H , Sukerobai M , Wang R . The Metabolic Syndrome in Uygur and Kazak Population. Diabetes Care . 28(10):2554-5, 2005.
- Zhou X, Huang J, Chen J, Zhao J, Ge D , Yang W, Gu D. Genetic association analysis of myocardial infarction with thrombospondin-1 N700S variant in a Chinese population. Thrombosis Research . 2004;113(3-4):181-186.
- Gu D, Ge D, He J, Li B, Chen J, Liu D, Chen J, Chen R. Haplotypic analyses of the aldosterone synthase gene CYP11B2 associated with stage-2 hypertension in northern Han Chinese. Clinical Genetics . 2004;66(5):409-416.
- Li B, Ge D, Wang Y, Zhao W, Zhou X, Gu D, Chen R. Lipoprotein lipase gene polymorphisms and blood pressure levels in the Northern Chinese Han population. Hypertension Research . 2004;27(6):373-378.
- Gu F, Ge D, Huang J, Chen J, Yang W, Gu D. Genetic susceptibility loci for essential hypertension and blood pressure on chromosome 17 in 147 Chinese pedigrees. Journal of Hypertension . 2004;22(8):1511-1518.
- Gu D, Ge D. The New Genetics in Hypertension: Asia Pacific Perspective. Paper presented at: 3rd Asian-Pacific Congress of Hypertension; April 3-7, 2004; Singapore.
W. Zeng
- Biotin-responsive basal ganglia disease maps to 2q36.3 and is due to mutations in SLC19A3. Zeng WQ, Al-Yamani E, Acierno JS Jr, Slaugenhaupt S, Gillis T, MacDonald ME, Ozand PT, Gusella JF. Am J Hum Genet. 2005 Jul;77(1):16-26. Impact Factor 12. 649
- Congenital lethal motor neuron disease with a novel defect in ribosome biogenesis. Butterfield RJ1, Stevenson TJ, Xing L, Newcomb TM, Nelson B, Zeng W, Li X, Lu HM, Lu H, Farwell Gonzalez KD, Wei JP, Chao EC, Prior TW, Snyder PJ,Bonkowsky JL, Swoboda KJ. Neurology. 2014 Apr 15;82(15):1322-30. Epub 2014 Mar 19. Impact Factor 8.3
- Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Farwell KD1, Shahmirzadi L1, El-Khechen D1, Powis Z1, Chao EC2, Davis BT1, Baxter RM1, Zeng W1, Mroske C1, Parra MC1, Gandomi SK1, Lu I1, Li X1, Lu H1, Lu HM1, Salvador D1, Ruble D1, Lao M1, Fischbach S1, Wen J1, Lee S1, Elliott A1, Dunlop CL1, Tang S1. Genetics in Medicine Impact Factor 6.44
- ELP2 is a novel gene implicated in neurodevelopmental disabilities. Cohen JS1, Srivastava S, Farwell KD, Lu HM, Zeng W, Lu H, Chao EC, Fatemi A. Am J Med Genet A. 2015 Apr 2.
- Diagnostic Exome Sequencing and Tailored Bioinformatics of the Parents of a Deceased Child with Cobalamin Deficiency Suggests Digenic Inheritance of the MTR and LMBRD1 Genes Farwell Gonzalez KD1, Li X, Lu HM, Lu H, Pellegrino JE, Miller RT, Zeng W, Chao EC. JIMD Rep. 2015;15:29-37. Epub 2014 Mar 25.
- Expanding the clinical and mutational spectrum of Kaufman oculocerebrofacial syndrome with biallelic UBE3B mutations. Basel-Vanagaite L1, Yilmaz R, Tang S, Reuter MS, Rahner N, Grange DK, Mortenson M, Koty P, Feenstra H, Farwell Gonzalez KD, Sticht H, Boddaert N,Désir J, Anyane-Yeboa K, Zweier C, Reis A, Kubisch C, Jewett T, Zeng W, Borck G. Hum Genet. 2014 Jul;133(7):939-49. Epub 2014 Mar 11.
- Xiang, B Xu, F., Zeng WQ, Dan Zi, Ma DQ, Navigating Web-Based Resources for Genetic Testing of Chromosome Abnormalities, CNVs and Gene Mutations. N A J Med Sci. 2014;7(4):163-170. DOI: 10.7156/najms. 2014.0704163
- Diagnostic Exome Sequencing Identifies Two Novel IQSEC2 Mutations Associated with X-Linked Intellectual Disability with Seizures: Implications for Genetic Counseling and Clinical Diagnosis. Gandomi SK, Farwell Gonzalez KD, Parra M, Shahmirzadi L, Mancuso J, Pichurin P, Temme R, Dugan S, Zeng W, Tang S. J Genet Couns. 2013.
- A human de novo mutation in MYH10 phenocopies the loss of function mutation in mice Rare Diseases 1. Tuzovic L, Yu L, Zeng W, Li X, Lu H, Lu HM, Gonzalez K, Chung WK. Rare Dis. 2013 Aug 14;1:e26144.
- Clinical exome sequencing leads to the diagnosis of mitochondrial complex I deficiency in a family with global developmental delays, ataxia, and cerebellar and pons hypoplasia Virginia Kimonis, Kelly Gonzalez, Wenqi Zeng, Phillip Gray, Sha Tang, Jennifer Wei, X. Li, HM. Lu, H. M. Lu, Elizabeth Chao Mitochondrion 11/2013; 13(6):942
- Zeng WQ, Gao H, Brueton L, Hutchin T, Gray G, Chakrapani A, Olpin S, Shih VE. Fumarase deficiency caused by homozygous P131R mutation and paternal partial isodisomy of chromosome 1. Am J Med Genet A. 2006; 140(9):1004-9. IF 2.44.
- Zeng W, Gillis T, Hakky M, Djoussé L, Myers RH, MacDonald ME, Gusella JF. Genetic analysis of the GRIK2 modifier effect in Huntington's disease. BMC Neurosci. 2006; 7;7:62 IF 2.987.
- Yan J, Noltner K, Feng J, Li W, Schroer R, Skinner C, Zeng W, Schwartz CE, Sommer SS. Neurexin 1alpha structural variants associated with autism. Neurosci Lett. 2008; 438(3):368-70. Epub 2008 Apr 25 IF 2.085.
- Zhao J, Chen Y, Zeng W et. Al. Pharmacokinetic and pharmacodynamics study of new antidepressant drug: Moclobemide Chinese Journal of Nervous and Mental Diseases. 中国神经精神疾病杂志, 1998; 24(8) 22-28.
- Zeng W, Zhao J. A new MAOI antidepressant drug: Moclobemide. International Journal of Psychiatry 国际精神病学杂志, 1995; 22 (1): 8-11.
- Zhang Y, Zeng W. A compared study of the copying style of different people that suffered from the different stress. 中国临床心理学杂志, 1994; 1(1): 6-10.
- Zeng W The Social Culture Explanation for Symptoms of Mental Disorders. Medicine and Philosophy医学与哲学,1994;15(11):5-8.
X. Guo
- X Guo, A Bernard, DA Orlando, SB Haase, & AJ Hartemink. (2013). “Branching process deconvolution algorithm reveals a detailed cell-cycle transcription program”, Proceedings of the National Academy of Sciences (PNAS), 110, E968-E977.
- MB Mayhew, X Guo, SB Haase, & AJ Hartemink. (2012). “Close encounters of the collaborative kind”, IEEE Computer, 45, pp. 24-30. (cover feature)
- X Guo, ML Bulyk, & AJ Hartemink. (2012). “Intrinsic disorder within and flanking the DNA-binding domains of human transcription factors”, In Pac Symp Biocomput 2012 (PSB12), pp. 104-115.
- X Guo, & AJ Hartemink. (2009). “Domain-oriented edge-based alignment of protein interaction networks”, Bioinformatics, 25, pp. i240-246.