Background
According to data from IARC, 19.29 million new cancer cases globally occurred in 2021, with about 9.96 million deaths. The global cancer burden is expected to reach 28.4 million cases in 2040, reflecting a 47% increase from 2020. Additionally, the five-year survival rates of cancer patients vary among different countries. The reasons for this are multifaceted. On one hand, the majority of cancers are commonly detected at a later stage, with most patients diagnosed at an intermediate or advanced stages, resulting in poor intervention outcomes. On the other hand, traditional early screening methods for cancer have many limitations, and for most types of cancer, effective early screening methods are still lacking, leading to generally low compliance. In recent years, with the advancement of molecular diagnostic techniques, liquid biopsy based on the methylation of circulating tumor DNA (ctDNA) has been a significant breakthrough in early cancer screening, demonstrating extensive potential applications. Currently, the methylation detection methods utilized for early cancer screening primarily include methylation-specific PCR (MSP), multiplex PCR amplification sequencing (Amp), and targeted methylation sequencing based on liquid-phase hybridization capture technology (Capture). MSP is only capable of qualitative detection of methylation and is generally used in the development of early screening products for a single type of cancer. Multiplex PCR amplification is applicable in the development of products for a single or multiple types of cancer, but its primer design is challenging. Apostle has launched the Apostle MiniEnrich targeted methylation sequencing solution, featuring an exclusive patented Apostle MiniEnrich liquid-phase hybridization capture system capable of completing the entire process within a single day, offering a simple and convenient operation. This solution captures the converted methylation libraries and uses a patented probe design scheme that covers all methylation states of the targets. Additionally, the hybridization capture system optimized for methylation is not only suitable for targeted enrichment with mini-Panels but also applicable for capturing special regions like AT-rich sequences.
Introduction
|
|
Introduction
Apostle MiniEnrich Targeted Methylation Sequencing Comprehensive Solution, featuring an exclusive patented Apostle MiniEnrich hybridization capture system, enabling the completions of the entire process within same day with simple and convenient operations. This solution captures the converted methylation libraries using a patented probe design scheme that covers targets in all methylation states. Furthermore, with a hybridization capture system optimized for methylation is not only suitable for targeted enrichment with mini-Panels but also applicable for capturing special regions like AT-rich sequences.
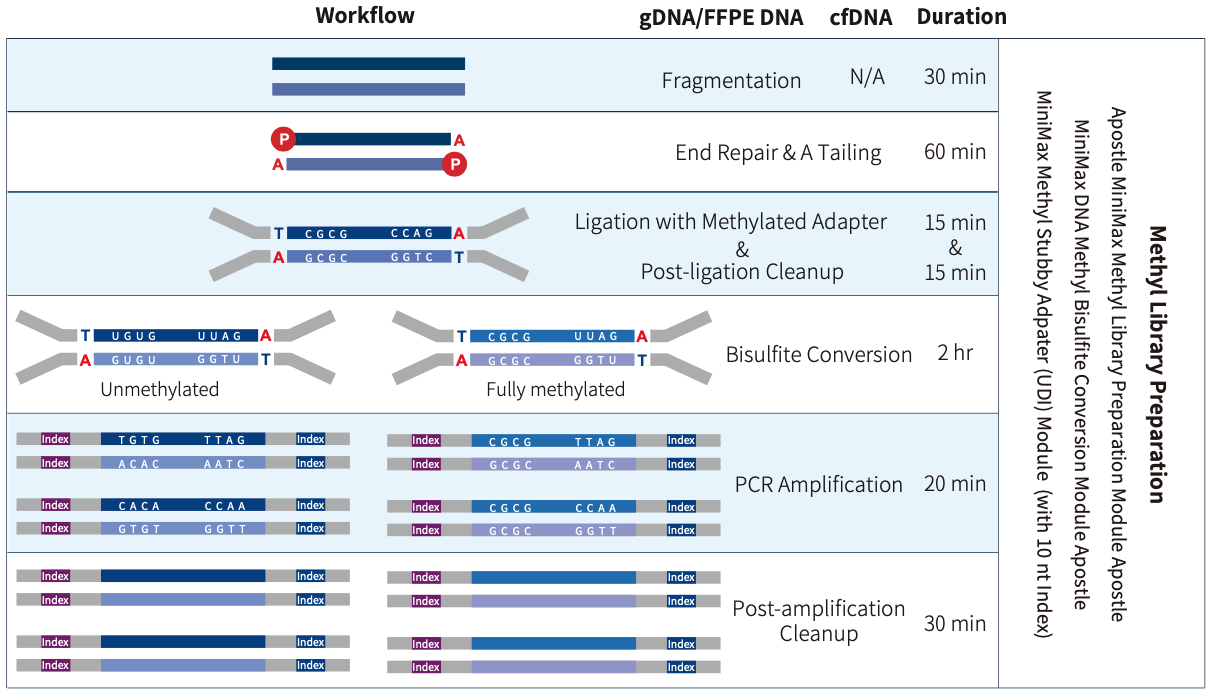
Apostle MiniEnrich Methyl Library Preparation Module is a double-strand DNA methylation sequencing library preparation kit developed for MGI & Illumina sequencing platforms. It supports multiple options for adapters selection, including Vendor N Methyl Adapter (SI) Module (for MGI), Vendor N Methyl Adapter (MDI) Module (for MGI) and Apostle MiniEnrich Methyl Stubby Adapter (UDI) Module (with 10 nt Index), etc. This kit can be used to prepare whole-genome methylation sequencing library. When combined with Apostle's liquid-phase hybridization capture series products, it enables targeted enrichment of methylation sequencing library.
Apostle MiniEnrich DNA Methyl Bisulfite Conversion Module is a methylation conversion reagent developed for DNA samples based on the principle of bisulfite conversion. The conversion module, based on a magnetic bead purification protocol, efficiently converts unmethylated cytosine to uracil within 2 hr, leaving methylated cytosine unchanged, and it can be compatible with automated workstations. It can be seamlessly integrated with Apostle's double-strand methylation library preparation scheme and single-strand methylation library preparation scheme, allowing flexible selection among various types of starting samples.
Apostle MiniEnrich Hybrid Capture Reagents v2 is designed for targeted enrichment of small Panel and hybrid capture of various types of pre-libraries, integrated with upgraded and optimized hybrid capture and elution processes, and equipped with Apostle MiniEnrich Panel designed based on innovative protocols, which can complete the whole process of capture-library preparation in same day.
Apostle MiniEnrich NanoBlockers are optimized blockers for MGI/Illumina platforms based on Apostle MiniEnrich Hybrid Capture System. The Apostle MiniEnrich NanoBlockers facilitates better binding of the library's adapter sequences to the sequencing platform. This reduces non-specific binding between adapters, resulting in improved on-target rates and increased data utilization. Apostle MiniEnrich NanoBlockers can be used to block the adapters with 6-10 nt index in the library.
Apostle MiniEnrich Panel is a commercial or customized Panel (Apostle MiniEnrich Custom Panel) based on Apostle's independent intellectual property rights with innovative scheme design. The panel can quickly find the binding position in the reaction system and further stabilize after multiple probes bind to the target region. If you hope to design personalized probes, please contact us (support@apostlebio.com) to get professional and personalized advice.
Feature
o Precise detection of large-scale CpG sites: Simultaneously assessment of multiple genes' CpG sites associated with various cancers, achieving precise comprehensive joint testing
o Accurate quantification of methylation levels: High-precision coverage for accurate quantification of different methylation states ranging from 0 to 100% in various samples
o Fast and convenient operation: Simplified experimental procedures, and user-friendly operations, completing the entire process within same day
o Highly efficient and stable: Avoid inconsistent amount of raw data, ensuring stability in capture sequencing results, thereby reducing the risk of rework
o Cost-effective sequencing: Utilize a mixed hybrid mode to reduce the quantity of hybrid reaction, lower sequencing costs and enhance cost-effectiveness
Performance
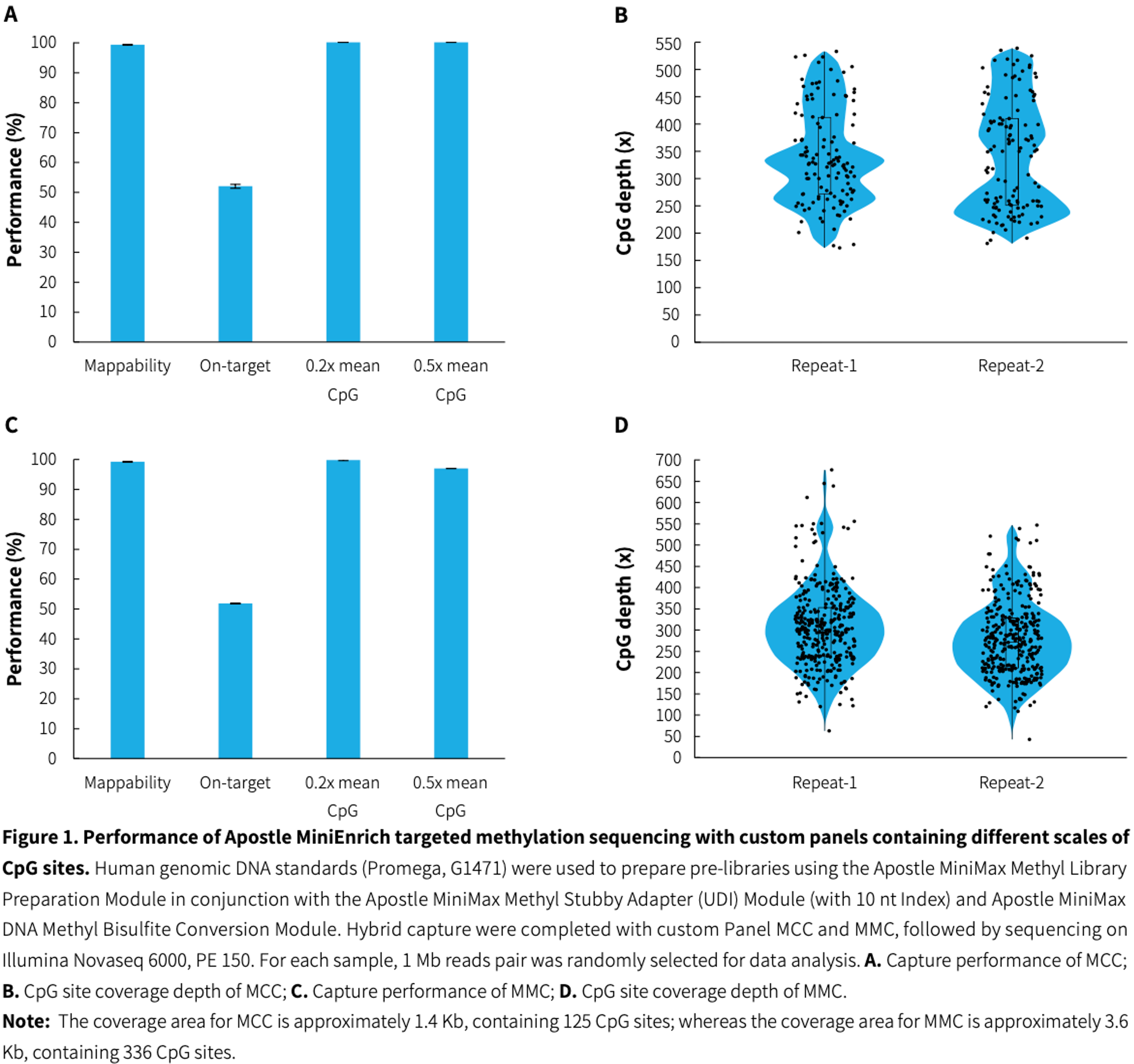
Precise detection of large-scale CpG sites
|
|
Accurate quantification of methylation levels
|
|
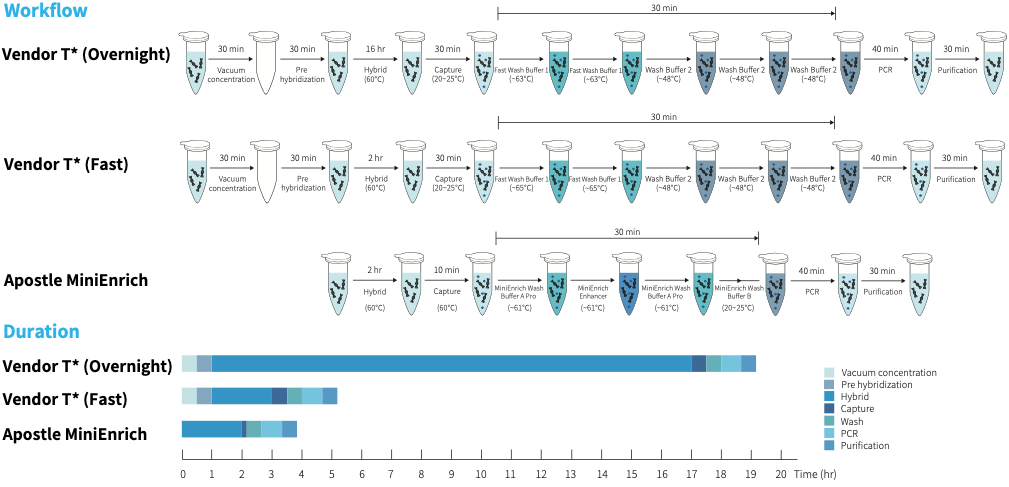
Fast and convenient operation
|
|
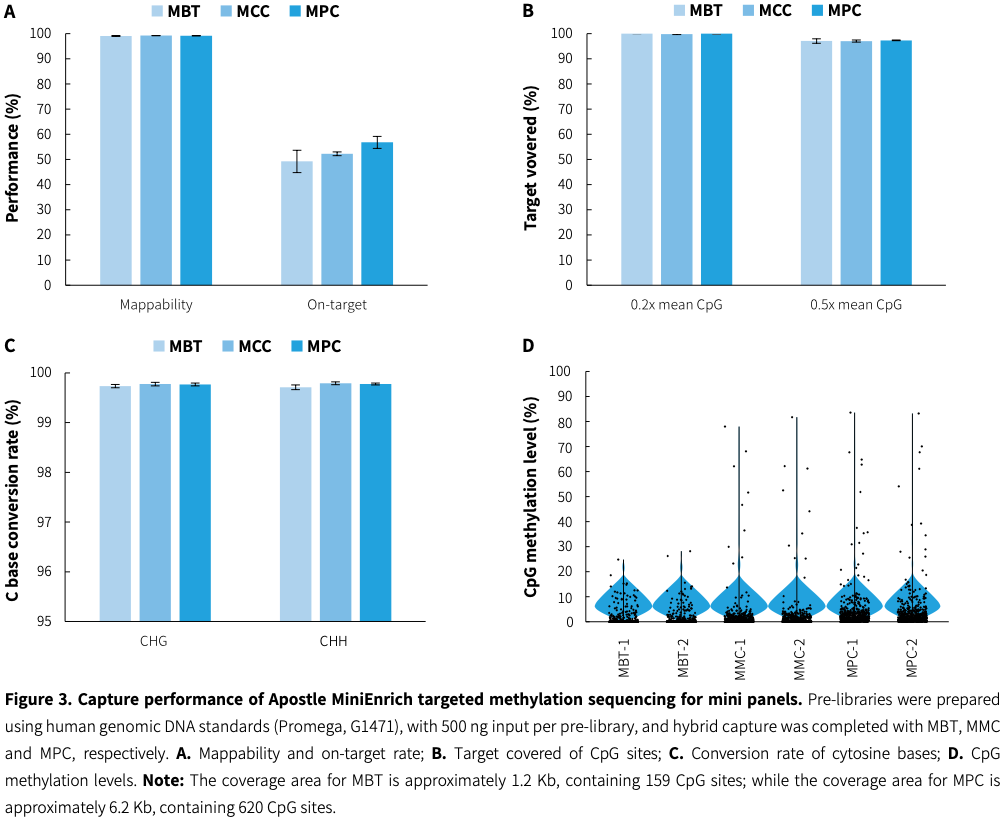
Highly efficient and stable
|
|
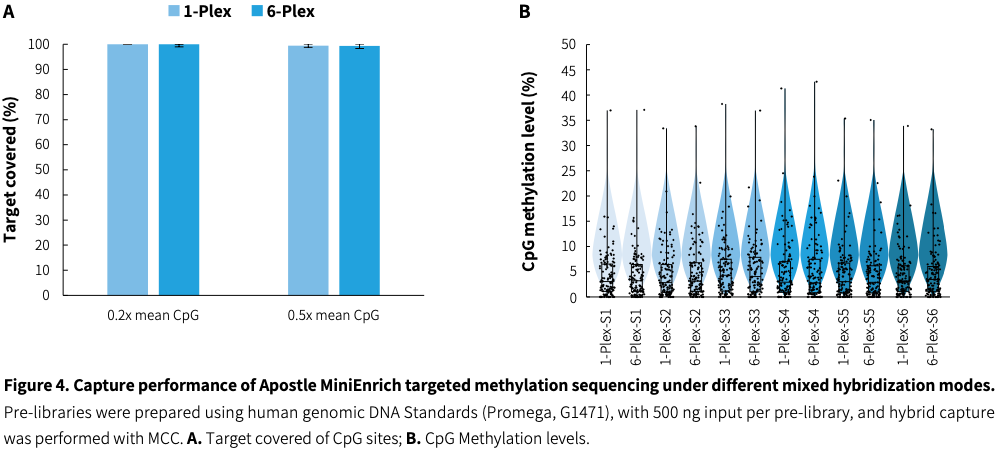
Cost-effective sequencing
|
|
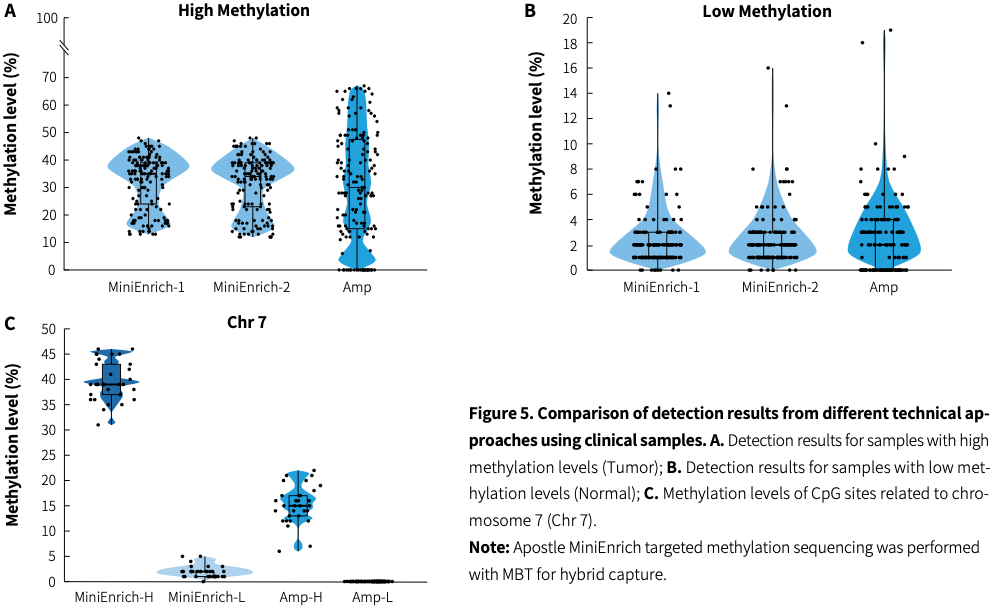
Application example of clinical sample
In cancer early screening cohort studies, various technical approaches are selected with the aim of validating methylation biomarkers screened across the whole genome, striving to meet multiple requirements such as high sensitivity, broad coverage, and cost-effectiveness. Researchers used Apostle MiniEnrich targeted methylation sequencing (MiniEnrich) and multiplex PCR amplification (Amp) to detect methylation levels of CpG sites in clinical gDNA samples with two different methylation levels.|
|